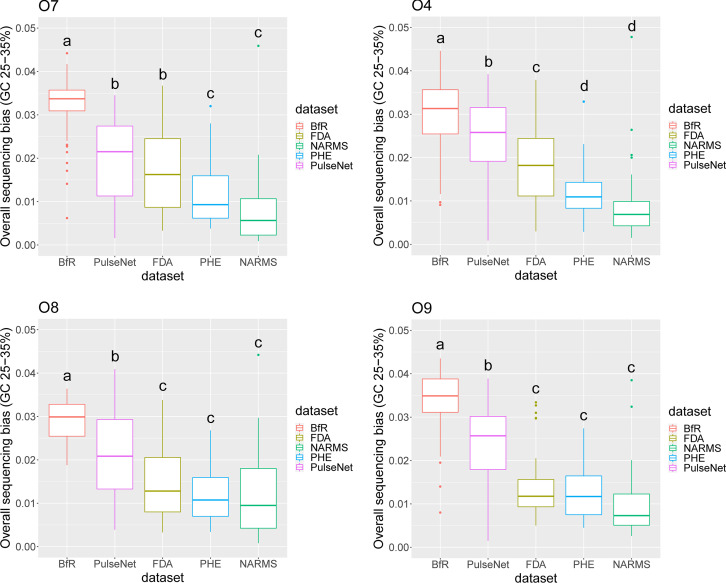
FIG 1.
Comparison of GC-associated sequencing biases of low-GC regions in genomes from four major O antigen groups. Low-GC regions are defined as regions of GC content between 25% and 35%, which is similar to the GC content of O antigen determinant genes (wzx/wzy). GC-associated sequencing bias was calculated according to methods described in reference 3. Salmonella genomes from five data sets were analyzed, including BfR (n = 415), PulseNet (n = 158), FDA (n = 145), Public Health England (PHE) (n = 160), and NARMS (n = 155). Genomes of four major O antigen groups (O7, O4, O8, and O9) in these data sets were analyzed. For the PulseNet, FDA, PHE, and NARMS data sets, the O7 group includes serotypes Braenderup, Infantis, Montevideo, and Thompson; the O4 group includes serotypes Agona, Heidelberg, Saintpaul and Typhimurium; the O8 group includes serotypes Hadar, Kentucky, Muenchen, and Newport; and the O9 group includes serotypes Berta, Enteritidis, Javiana, and Panama. Up to 10 genomes of each serotype were randomly selected from each data set. For Nextera XT-prepared genomes in the study by Uelze et al. (n = 578), all the O7, O4, O8, and O9 genomes were included except serotype Enteritidis. This serotype was overrepresented in that study (n = 115), and 20 genomes were randomly selected for this analysis. Different letters above boxes indicate that there is a significant difference (P < 0.05, analysis of variance [ANOVA]) between the values.