In this study, AOA communities along a river in southern China were characterized, and metagenome-assembled genomes (MAGs) of a novel AOA clade were also obtained. Based on the characterization of AOA genomes, the study suggests adaptation of the novel AOAs to estuarine environments, providing new information on the ecology of estuarine AOA and the nitrogen cycle in contaminated estuarine environments.
KEYWORDS: Jiulong River estuary, nitrification, ammonia-oxidizing archaeon, metagenome
ABSTRACT
Ammonia-oxidizing archaea (AOA) are ubiquitous in diverse ecosystems and play a pivotal role in global nitrogen and carbon cycling. Although AOA diversity and distribution are widely studied, mainly based on the amoA (alpha subunit of ammonia monooxygenase) genotypes, only limited investigations have addressed the relationship between AOA genetic adaptation, metabolic features, and ecological niches, especially in estuaries. Here, we describe the AOA communities along the Jiulong River estuary in southern China. Nine high-quality AOA metagenome-assembled genomes (MAGs) were obtained by metagenomics. Five of the MAGs are proposed to constitute a new species, “Candidatus Nitrosopumilus aestuariumsis” sp. nov., based on the phylogenies of the 16S and 23S rRNA genes and concatenated ribosomal proteins, as well as the average amino acid identity. Comparative genomic analysis revealed unique features of the new species, including a high number of genes related to diverse carbohydrate-active enzymes, phosphatases, heavy-metal transport systems, flagellation, and chemotaxis. These genes may be crucial for AOA adaptation to the eutrophic and heavy-metal-contaminated Jiulong River estuary. The uncovered detailed genomic characteristics of the new estuarine AOA species highlight AOA contributions to ammonia oxidation in the Jiulong River estuary.
IMPORTANCE In this study, AOA communities along a river in southern China were characterized, and metagenome-assembled genomes (MAGs) of a novel AOA clade were also obtained. Based on the characterization of AOA genomes, the study suggests adaptation of the novel AOAs to estuarine environments, providing new information on the ecology of estuarine AOA and the nitrogen cycle in contaminated estuarine environments.
INTRODUCTION
Microbial nitrification is an important and regulatory process in the terrestrial and aquatic nitrogen cycles. Ammonia oxidation is the first, and rate-limiting, metabolic reaction catalyzed by ammonia-oxidizing bacteria (AOB) (1), ammonia-oxidizing archaea (AOA) (2, 3), and complete ammonia oxidizers (comammox bacteria) (4, 5).
AOA are ubiquitous and abundant in the terrestrial and marine biosphere, including the soil (6) and oceanic water and sediment (3, 7), and in acidic and geothermal habitats (8, 9). According to previous surveys, AOA distribution is environment driven, which leads to the formation of different ecotypes with various metabolic capacities (10–12).
To date, AOA (class Nitrososphaeria) consist of four identified orders, i.e., Nitrosopumilales (MG-1; group 1.1a), “Candidatus Nitrosotaleales” (SAGMCG-1; group 1.1a associated), Nitrososphaerales (SCG; group 1.1b), and “Candidatus Nitrosocaldales” (HWCGIII; ThAOA) (13). Nitrosopumilus-like AOA are the most abundant AOA in shallow water and surface sediments, especially in estuarine and coastal regions (13–15). However, the phylogeny of the 16S rRNA genes and amoA genes indicates that the diversity of AOA exceeds the current understanding (10, 16). The ever-increasing amount of genomic information on novel types of AOA illustrates their ecological roles and metabolic functions.
Following the initial discovery of Nitrosopumilus maritimus strain SCM1 (SCM1) (2), AOA have been isolated or enriched from diverse environments, suggesting that the ability to adapt is one of the crucial characteristics underpinning the ubiquitous AOA distribution. Some examples are mesophilic and neutrophilic “Candidatus Nitrosomarinus catalina” SPOT01 (SPOT01) (17) from open-ocean waters, “Candidatus Nitrosocosmicus oleophilus” MY3 (18) from a coal tar-contaminated sediment, and Nitrososphaera viennensis EN76 (19) from garden soil; acidophilic Nitrosopumilus ureiphilus PS0 (20) and “Candidatus Nitrosotalea devanaterra” Nd1 (21), isolated from coastal water and soil, respectively; thermophilic “Candidatus Nitrosocaldus islandicus” (22), and “Candidatus Nitrosocaldus cavascurensis” (23), both from hot springs; and “Candidatus Nitrosopumilus salaria” BD31 (24) and “Candidatus Nitrosoarchaeum limnia” SFB1 (25), isolated from low-salinity estuarine sediments. Although knowledge of AOA physiology has been largely supported by cultivation experiments, most lineages remain uncultured. Therefore, detailed genomic mechanisms underpinning adaptation to distinct environments are pivotal to improving our understanding of AOA universality and diversification.
The Jiulong River estuary is located in southeast China. Pronounced nitrification and denitrification processes have been reported in the estuary (26, 27). Although relatively high microbial activity related to nitrogen metabolism in the area, including AOA (28, 29), was reported in several studies, the distribution and genomic adaptations of AOA there are poorly understood. In the current study, we quantified the abundance of AOA and AOB amoA genes in surface waters and sediments in the Jiulong River estuary. We observed that the ratio of AOA over AOB increased with increasing salinity. Using metagenomic analysis, we categorized the genotypes of identified archaeal amoA genes. Further, we reconstructed nine nearly complete AOA metagenome-assembled genomes (MAGs) from the water and sediment samples. Notably, five AOA MAGs constituted a potential new species within the genus Nitrosopumilus, which was supported by phylogenetic analysis of concatenated ribosomal proteins, average amino acid identity (AAI), and 16S rRNA gene similarity analyses; the species was named “Candidatus Nitrosopumilus aestuariumsis” sp. nov., according to recently proposed roadmaps for nomenclature of uncultivated archaea (30). Genomic and pangenomic analyses revealed that the novel AOA species carries a high number of genes related to carbohydrate metabolism, transport systems, and chemotaxic proteins. We also demonstrated clear community compositions and variations of AOA in the study area and revealed specific genetic features associated with the different adaptations.
RESULTS
Geographic locations, environmental variables, and nitrification rates.
The concentrations of inorganic nutrients (NO2−, NO3−, NH4+, and PO43−) and environmental parameters (dissolved oxygen [DO], salinity, and pH) at all sampling sites are presented in Table 1. The salinity of the surface water increased from 0.5‰ in water sample W1 to 27.0‰ in water sample W3. The concentration of inorganic nutrients determined in the current study decreased gradually from W1 to W3, with the salinity increasing along the estuary from head to mouth. The highest nitrification rate was detected at W1 and was reduced to the lowest at W3. DO concentrations were much lower in the bottom water (1.35 to 2.03 mg/liter) than in the surface water (2.46 to 7.46 mg/liter), which indicated a microaerobic condition in the former at the time of sampling.
TABLE 1.
Summary information for sampling sites
| Station | Longitude [°E] | Latitude [°N] | Water depth (m) | NO2− (μM) | NO3− (μM) | NH4+ (μM) | DO (mg/liter) | PO43− (μM) | Salinity (‰) | pH | Chl a (μg/liter) | Ammonia oxidation rate (nM/h)a |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Shallow-water samples | ||||||||||||
| W1 | 117.79 | 24.47 | 2.0 | 13.30 | 214.8 | 35.18 | 2.46 | 2.07 | 0.5 | 7.23 | 11.49 | 81.57 ± 8.69 |
| W2 | 117.97 | 24.40 | 2.0 | 12.85 | 99.1 | 12.83 | 6.73 | 1.92 | 15.8 | 7.56 | 5.85 | 10.25 ± 0.12 |
| W3 | 118.11 | 24.38 | 2.0 | 6.23 | 43.1 | 2.68 | 7.46 | 1.25 | 27.0 | 7.81 | 5.22 | 9.88 ± 0.23 |
| Sediment samplesb | ||||||||||||
| S1 | 117.79 | 24.47 | 7.0 | 10.41 | 203.6 | 19.69 | 1.93 | 1.66 | 0.1 | 7.39 | 10.73 | |
| S2 | 117.92 | 24.39 | 6.0 | 12.73 | 132.1 | 15.61 | 2.03 | 0.83 | 12.3 | 7.45 | 6.23 | |
| S3 | 113.105 | 24.42 | 10.3 | 6.81 | 53.8 | 3.00 | 1.35 | 3.19 | 26.7 | 7.76 | 5.7 |
The rate was not determined for bottom-water samples.
The environmental factor measured was the bottom water above each sediment sample.
Archaeal abundance and community composition.
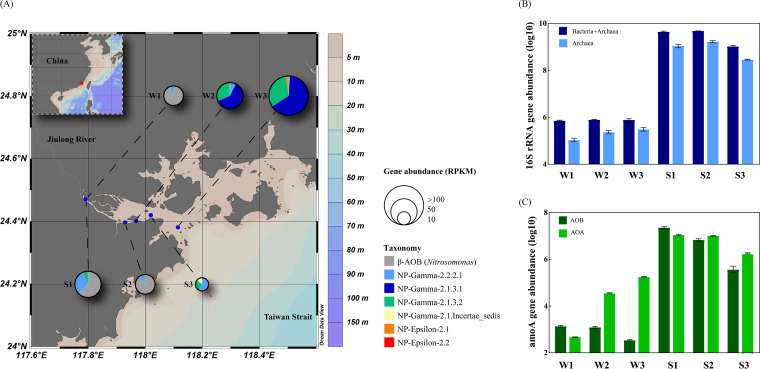
The copy numbers of all prokaryotic (bacterial and archaeal) 16S rRNA genes were approximately 7.25 × 105 to 7.91 × 105 copies/ml in water samples and approximately 1.05 × 109 to 4.80 × 109 copies/g in sediment samples (Fig. 1B; see Table S2 in the supplemental material). The abundance of archaeal 16S rRNA genes increased from approximately 1.41 × 105 copies/ml in low-salinity water sample W1 to approximately 3.09 × 105 copies/ml in high-salinity water sample W3, accounting for 19.4% to 39.3% of all prokaryotes. In sediment samples, the abundance of archaeal 16S rRNA genes was lowest in sample S3 (ca. 3.72 × 108 copies/g) and highest in sample S2 (ca. 2.14 × 109 copies/g), and archaea accounted for 31.4% to 44.6% of all prokaryotes. Gel electrophoretograms for each primer pair (see Fig. S1 in the supplemental material) and melting curve plots for each real-time quantitative-PCR (qPCR) experiment (see Fig. S2 in the supplemental material) indicated that the results were reliable.
FIG 1.
(A) Relative abundances and community compositions of amoA genes at the sampling stations based on metagenomic sequences. (B and C) Quantified abundances (log10) of total bacterial and archaeal 16S rRNA genes (B) and bacterial and archaeal amoA genes (C) in samples. The maps were created using Ocean Data View software.
The N50 for the total assembled scaffolds for each sample exceeded 1,350 bp, indicating the high quality of the assembly (see Table S3 in the supplemental material). All the bacterial and archaeal 16S rRNA genes were extracted from the assembled scaffolds to calculate the relative abundance (see Fig. S3A in the supplemental material), and Bathyarchaeota, Euryarchaeota, Woesearchaeota, and Thaumarchaeota were the major archaeal phyla identified in the current study (see Fig. S3B). Further, the abundance of Thaumarchaeota in water samples increased with increasing salinity, but it did not follow any clear pattern in sediment samples. All the MAGs were of relatively high quality (completeness, >70%, and contamination, <10%) (Table 2).
TABLE 2.
Basic information for reconstructed AOA MAGs
| Bin | Size (Mb) | Completeness (%) | Contamination (%) | No. of genesa |
|---|---|---|---|---|
| S1bin1 | 1.31 | 91.64 | 1.94 | 1,698 |
| S2bin1 | 1.44 | 95.63 | 0.97 | 1,861 |
| S3bin1 | 1.24 | 89.81 | 3.87 | 1,669 |
| W1bin1 | 1.40 | 91.75 | 4.37 | 1,795 |
| W2bin1 | 0.85 | 72.98 | 5.83 | 1,145 |
| W2bin2 | 1.13 | 89.32 | 4.85 | 1,530 |
| W2bin3 | 1.28 | 78.02 | 3.88 | 1,796 |
| W3bin1 | 1.26 | 64.89 | 5.34 | 1,779 |
| W3bin2 | 0.88 | 79.29 | 7.77 | 1,152 |
Genes were predicted using Prodigal (version 2.6.3).
AOA abundance, diversity, and distribution.
Notably, the archaeal amoA gene copy number dramatically increased from the low-salinity sample W1 (ca. 4.69 × 102 copies/ml) to the high-salinity sample W3 (ca. 1.81 × 105 copies/ml), while the abundance of bacterial amoA genes decreased with increasing salinity, from approximately 1.36 × 106 copies/liter to approximately 3.46 × 105 copies/liter (Fig. 1C; see Table S2). In contrast, the abundances of both bacterial and archaeal amoA genes were highest in the sediment sample S1 (ca. 2.27 × 107 copies/g and ca. 1.10 × 107 copies/g, respectively) and gradually decreased along the salinity gradient. The abundance of archaeal amoA genes exceeded that of bacterial genes in sample S3 (ca. 1.80 × 106 copies/g versus ca. 4.62 × 105 copies/g, respectively) (Fig. 1C; see Table S2). Similar patterns were apparent in the metagenomic data, in that the relative abundance of amoA genes increased along the estuary in water samples but decreased in sediment samples, and archaeal amoA genes dominated in high-salinity regions (Fig. 1A).
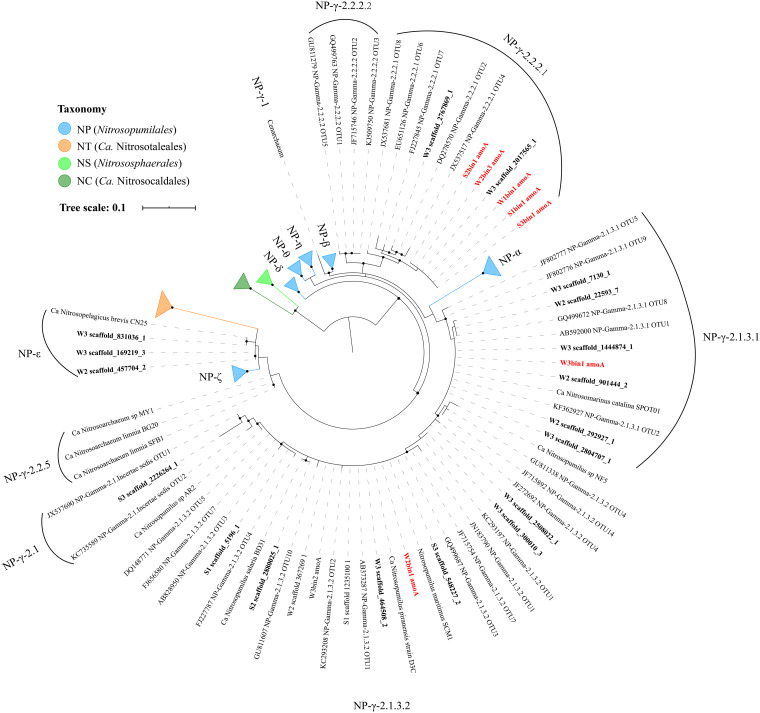
All the bacterial amoA genes identified in the current study were from β-AOB (genus Nitrosomonas), while all archaeal amoA genes belonged to the order Nitrosopumilales (NP) and were classified into several subgroups defined by Alves et al. (16), i.e., NP-γ (Nitrosopumilus SCM1-like group [SCM1-like]) and NP-ε (water column A group [WCA]) (Fig. 2). Both NP-γ-2.1.3.2 and NP-γ-2.2.2.1 genotypes were commonly categorized in the sediment and water samples, while genes within NP-γ-2.1.Incertae_sedis and NP-γ-2.1.3.1 were identified exclusively in the sediment and water samples, respectively. Notably, amoA genes affiliated with NP-γ-2.2.2.1 were observed in all the samples, i.e., in five MAGs (S1bin1, S2bin1, S3bin1, W1bin1, and W2bin3) and two unbinned sequences from W3 (Fig. 2 and 3). Along the salinity increase, more genotypes of archaeal amoA genes were observed (Fig. 1A). The predominant genotype of archaeal amoA genes in the water changed from NP-γ-2.2.2.1 to NP-γ-2.1.3.1, while the NP-γ-2.2.2.1 genotype was dominant in all the sediment samples.
FIG 2.
Phylogenetic tree of amoA genes extracted from AOA MAGs (boldface red) and unbinned metagenomic data for samples (boldface black), using an mtZOA+G4 model with 248 aligned amino acid residues. No amoA genes were identified in bins W2bin2 and W3bin1. Bootstrap values of >90% are denoted by solid circles.
FIG 3.
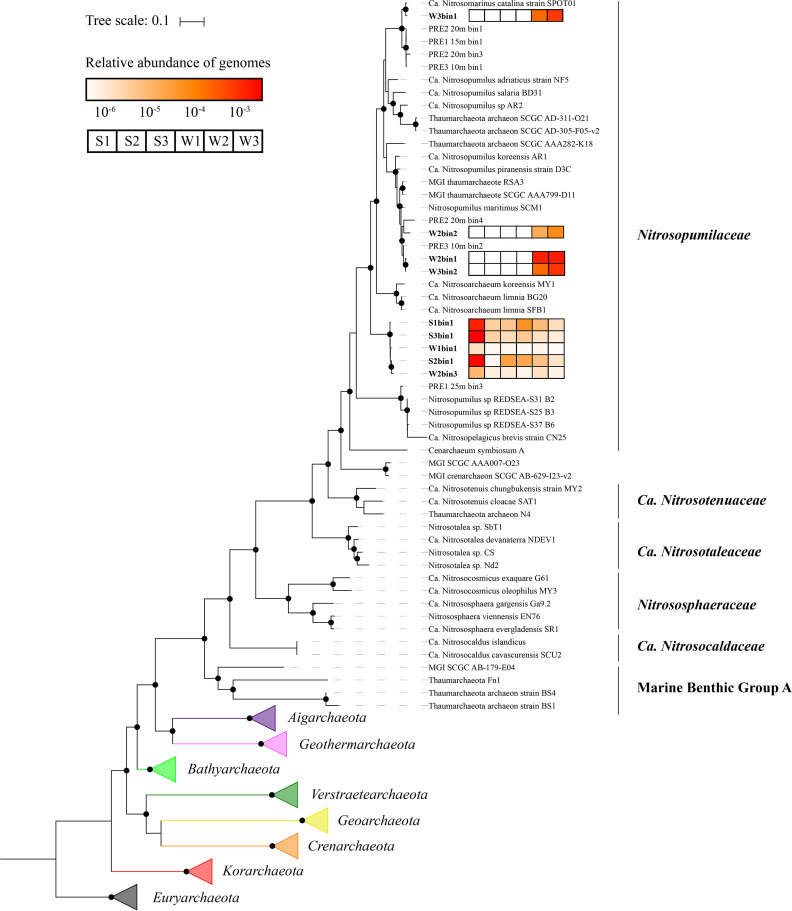
Phylogenetic tree of 15 concatenated ribosomal proteins for AOA MAGs (boldface), AOA reference genomes, and other archaea from the TACK superphylum. Euryarchaeotal genomes were used as the outgroup, using an LG+C60+F model with 4,680 aligned amino acid residues. Bootstrap values of >90% are denoted by solid circles. The heat map represents the relative abundance of each MAG in all samples.
All AOA MAGs reconstructed in the current study were identified based on the phylogeny of 15 concatenated ribosomal-protein sequences, including reference genomes of AOA isolates/enrichments and other reported MAGs (Fig. 3). W2bin1, W2bin2, and W3bin2 were affiliated with the marine AOA strain SCM1, while W3bin1 was located close to the coastal strain SPOT01 in the phylogenetic tree. Based on the relative abundance, the predominant AOA shifted to SCM1-like and SPOT-like in high-salinity regions, but these types were rarely detected in low-salinity samples. Intriguingly, five AOA MAGs (S1bin1, S2bin1, S3bin1, W1bin1, and W2bin3) were clustered together but not affiliated with any known “Candidatus” genera or species from the family Nitrosopumilales. According to the relative-abundance analysis, the novel AOA cluster may inhabit both aquatic and sedimentary environments along the estuary but be enriched in low-salinity regions.
Phylogenetic identification and genomic features of the novel AOA genus.
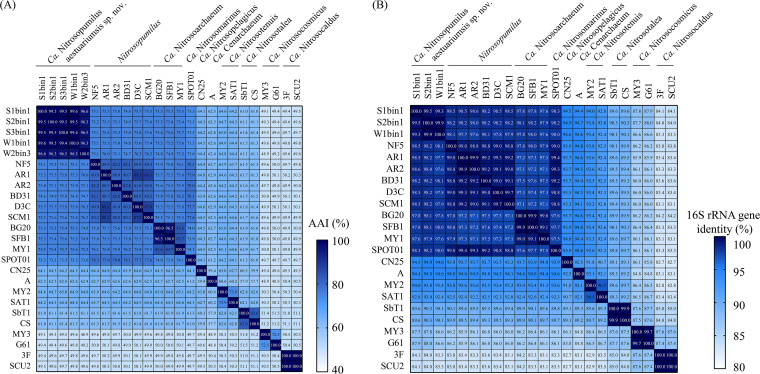
To confirm the taxonomy unit of the novel AOA cluster, the phylogeny and identity of 16S and 23S rRNA genes and AAI shared with different AOA species were used. The AAI among all MAGs in this cluster exceeded 96.3%, with >80.2% and >83.6% values for the genera Nitrosopumilus and “Candidatus Nitrosoarchaeum,” respectively (Fig. 4A). Further, the novel AOA cluster shared 74.6 to 76.3%, 74.8 to 75.7%, and 73.3 to 73.6% similarity with the genera Nitrosopumilus, “Ca. Nitrosoarchaeum,” and “Candidatus Nitrosomarinus,” respectively. These values are similar to the AAI values between the current “Candidatus” genera in the family Nitrosopumilaceae (i.e., >70%) (Fig. 4A). For the 16S rRNA genes, the novel AOA shared >97.6% similarity with other “Candidatus” genera in the family Nitrosopumilaceae, while these values were similar among other “Candidatus” genera (i.e., >97%) (Fig. 4B). Thus, according to previous studies (31, 32), these results suggested equal taxonomic levels for the AOA cluster and other “Candidatus” genera, and they should be reclassified as species level under the genus Nitrosopumilus based on the AAI and 16S rRNA gene similarity. In the phylogenetic tree of concatenated 16S and 23S rRNA genes, the novel AOA formed a cluster located separately in Nitrosopumilaceae (see Fig. S4 in the supplemental material). This was in agreement with the phylogeny based on ribosomal proteins and amoA genes (Fig. 2 and 3). Here, we name the novel species “Candidatus Nitrosopumilus aestuariumsis” sp. nov.
FIG 4.
Heat maps illustrating results for AAI (A) and 16S rRNA gene identity (B) analyses between AOA MAGs from the current study (i.e., S1bin1, S2bin1, S3bin1, W1bin1, and W2bin3) and other AOA isolates from different genera. No 16S rRNA genes were identified in bins S3bin1 and W2bin3.
Based on the reconstructed metabolic pathways, the novel AOA species encodes major carbon metabolic pathways commonly shared by other AOA, including a nearly complete gluconeogenesis pathway, a nonoxidative pentose-phosphate pathway, a tri-carboxylic acid cycle pathway, and a 3-hydroxypropionate/4-hydroxybutyrate pathway (see Fig. S5 in the supplemental material). Further, a nearly complete pathway for cobalamin (vitamin B12) biosynthesis was annotated in all the MAGs. Single copies of amoC and nirK genes, but no ureC gene, were also identified in the novel AOA genus. MAGs in the novel AOA genus also encoded an A-type ATPase, as determined by BLAST analysis of atp genes. The arrangement of eight A-type ATPase genes (encoding subunits F, E, A, B, D, K, I, and C) was consistent with those of other coastal and estuarine AOA isolates/enrichments, such as strain SPOT01. The novel AOA also encoded transporters for oligopeptides, dipeptides, and amino acids, as well as the NitT/TauT family transport system (for nitrate, sulfonate, and bicarbonate), the ABC-2-type transport system (for polysaccharides and polyol-phosphate), and fluoride ion and lipoprotein export systems. Heavy-metal transporters for magnesium, potassium, zinc, copper, nickel, cobalt, and iron were also identified. No high-affinity pst transporter genes (pstABCS) were apparent, while genes for the low-affinity pit transporter and phosphonate transporters (phnCDE) were widely carried. In addition, genes for polyphosphate utilization (ppA) were present, as well as 2 types of ammonium transporters for both high affinity (amt-1) and low affinity (amt-2) (33) in all MAGs.
Carbohydrate-active enzymes (CAZys) and extracellular peptidases were annotated in the novel genus and compared with those of other AOA isolates (see Tables S4 and S5 in the supplemental material). The major glycosyltransferases belonged to family 2 (GT2), which is commonly encoded by other AOA isolates; the enzymes include cellulose synthase, dolichyl-phosphate β-d-mannosyltransferase, N-acetylglucosaminyltransferase, and chitin oligosaccharide synthase. Glycosyltransferases from the families GT1 (UDP-glucuronosyltransferase) and GT66 (dolichyl-diphosphooligosaccharide glycotransferase) were also prevalent. Interestingly, genes encoding chitinase (glycoside hydrolase family 18 [GH18]) were identified in all novel AOA MAGs, but not in other Nitrosopumilaceae AOA. BLAST annotation revealed that these hydrolase genes were more similar to bacterial chitinase genes than to archaeal chitinase genes, which was confirmed by phylogenetic analysis (see Fig. S6 in the supplemental material). It is highly probable that the chitinase genes were transferred from bacteria via horizontal gene transfer. Genes for extracellular peptidases, including ggt (gamma-glutamyltranspeptidase/glutathione hydrolase) and aprE (serine protease from the peptidase S8 family), were identified, and genes encoding extracellular enzymes, such as superoxide dismutase and RNase, were also commonly observed.
Genomic comparisons and pangenomic analysis of different AOA species.
The cluster of orthologous genes (COG) function categories in the estuarine AOA gene pool and the marine AOA gene pool are compared in Fig. S7 and Table S6 in the supplemental material. Generally, genes carried only by the estuarine AOA represented 198 categories; those carried only by the marine AOA represented 71 categories. Estuarine AOA carried more unique genes for amino acid and lipid metabolism, signal transduction, cell mobility, and transport than marine AOA.
To better understand the unique genomic features of the novel AOA species from the Jiulong River estuary, genome comparisons against each AOA species were made based on the COG gene categories (see Fig. S8 and Table S7 in the supplemental material). No significant differences in the compositions of COG categories were apparent among them. However, detailed pairwise comparison revealed that the novel AOA species comprised 30, 40, and 82 unique COG gene categories compared with Nitrosopumilus, “Ca. Nitrosoarchaeum,” and “Ca. Nitrosomarinus,” respectively. Except for the genes with unknown function, the majority of unique genes were associated with amino acid and carbohydrate metabolism, inorganic ion transport, and signal transduction.
Genes from the AOA isolates and MAGs reconstructed in the current study were then clustered based on similarity. Subsequent pangenomic analysis revealed genes carried only by the novel AOA (see Fig. S9 in the supplemental material). Overall, 206 gene clusters were identified as uniquely carried by the novel AOA, with detailed annotation for only 50 (others mainly encoded hypothetical proteins with unknown functions [see Table S8 in the supplemental material]). Notably, the unique genes encoded a variety of enzymes related to carbohydrate metabolism, such as chitinase, polyketide cyclase, peptidoglycan/xylan/chitin deacetylase, 2-hydroxy-3-oxopropionate reductase, Zn-dependent protease, glyoxylase dehydrogenase, and arylsulfatase. Genes for phosphoserine phosphatase, phosphoglycolate phosphatase, and flavin mononucleotide phosphatase were also identified. In addition, genes related to chemotaxis and archaellum biogenesis, signal transduction, and DNA repair and recombination were prevalent in the novel species.
DISCUSSION
Shift of the ammonia oxidizer community along the Jiulong River estuary.
The rates of ammonia oxidation detected in the current study varied from approximately 10 nM/h to >80 nM/h (approximately 240 nM/day to >1,920 nM/day) in the surface water. These values were much higher than those reported for pelagic surface water in the Pacific (<25 nM/day) (34) and the Atlantic (<10 nM/day) (35). Also, the rates were comparable to those in other coastal and estuarine surface waters, such as the Pearl River estuary (36), the Changjiang River estuary (37), the San Francisco Bay estuary (38), the Cochin estuary (39), and the Ems estuary (40). Pronounced ammonia oxidation in the waters of the Jiulong River estuary indicated potentially higher activity of ammonia oxidizers therein. Although comammox bacteria were reported to be widely distributed in both man-made systems and natural environments, including terrestrial, coastal, and open-ocean sites (41–44), yet no amoA genes of the bacteria were detected in the current study based on metagenomics, suggesting that AOA and AOB were major ammonia oxidizers in the estuary. The abundances of both archaeal and bacterial amoA genes in the estuarine waters exceeded those reported for the Dongjiang River (45) and the Pearl River delta (46). The abundances of sedimentary amoA genes detected in the current study were higher than those in other subtropical estuarine sediments, such as the Pearl River estuary (47) and the San Francisco Bay estuary (48).
Salinity is one of the most important factors that shape the microbial community and affect microbial activities, especially in the estuarine ecosystem (49, 50). In the current study, the abundance and composition of ammonia oxidizers dramatically changed along the salinity gradient, with the abundance of AOA overwhelmingly greater than that of β-AOB in high-salinity samples (Fig. 1). β-AOB are widely distributed in diverse environments, and members of the genus Nitrosomonas are commonly observed in both terrestrial and marine environments (51–54), as well as in the drinking water system and wastewater treatment plants (55, 56). Based on culture experiments (57) and in situ estuarine samples (58–60), Nitrosomonas organisms are more abundant and active under low-salinity conditions than under high-salinity conditions. In contrast, AOA tend to tolerate a wide salinity range, which is their key characteristic as the most ubiquitous ammonia oxidizers globally (2, 16, 17, 25). Furthermore, AOA have a higher affinity for ammonia than AOB and thus can survive and outcompete them in extremely low ammonium concentrations (61). That is in agreement with the findings of the current study on AOA dominance with decreasing ammonium concentrations in the estuary (Table 1). The abundance of archaeal 16S and amoA genes increased with salinity in the surface water but decreased in the surface sediment (Fig. 1; see Table S1 in the supplemental material), which was analogous to the changes in archaeal and thaumarcheotal abundance fractions (Fig. S3A and B). Besides salinity, the abundance of AOA was reported to be positively correlated with pH and DO and ammonium concentrations in estuarine and coastal sediments (47, 62–65). The decreased DO and ammonium concentrations along the estuary may be responsible for the decreased abundance of AOA in this study.
Of note, the AOA diversity also increased from the estuary head to its mouth, based on the genotype of archaeal amoA genes (Fig. 1). The oceanic NP-γ-2.1.3.1 (SPOT01-like) and NP-γ-2.1.3.2 (SCM1-like) were the major amoA genotypes in intermediate- and high-salinity waters in the current study, which was in agreement with the findings for the Pearl River estuary (36). The AOA distribution reported here also indicated that the oceanic SPOT01-like and SCM1-like AOA were more abundant in high-salinity waters (Fig. 3). In contrast, the seldom-reported genotype NP-γ-2.2.2.1 was dominant in low-salinity waters and all sediment samples. The reference sequences for genotype NP-γ-2.2.2.1 in the phylogenetic tree shown in Fig. 2 were obtained from sediments in the South China Sea and the San Francisco Bay estuary. The identification of genotype NP-γ-2.2.2.1 amoA genes in low- and high-salinity waters offered additional information on the abundance and distribution of the genotype. Notably, the genotype NP-γ-2.2.2.1 AOA were widely distributed in the Jiulong River estuary, based on the distribution and relative abundance of both amoA genes and MAGs, implying their contributions to both aquatic and sedimentary ammonia oxidation and highlighting their unquestionably important role in the local ecosystem.
Genomic characteristics of the novel AOA species.
Phylogenetic analysis revealed that five MAGs representing the NP-γ-2.2.2.1 amoA genotype formed a unique clustered branch within the family Nitrosopumilaceae and were not affiliated with any known AOA species (Fig. 3; see Fig. S4). In the current study, we suggest a species level taxonomy unit for these AOA in the genus Nitrosopumilus. Although the distribution of NP-γ-2.2.2.1-type AOA has been mainly based on the analysis of amoA and 16S rRNA genes (48, 66), limited genomic information is available for this AOA group, as no cultured strains or MAGs have been reported. Here, we reconstructed five high-quality genomes potentially representing a new species for the NP-γ-2.2.2.1-type AOA and reported their detailed genomic characteristics and metabolic capacities.
According to the COG functional-category analysis, estuarine AOA harbor multiple genes related to metabolism, signaling, mobility, and transport, which may illustrate their strategy to thrive in the changeable estuarine environment. Based on the proposed thaumarchaeotal core genome (67), the major metabolic pathways common among AOA isolates from the family Nitrosopumilaceae, such as carbon fixation via the 3-hydroxypropionate/4-hydroxybutyrate cycle (68), were reconstructed in the current study (see Fig. S5). Environment-specific energy-yielding ATPases in Thaumarchaeota that use A-type ATPases are mainly identified in neutrophilic AOA from terrestrial, shallow marine, estuarine, and coastal environments (12). The observations on the AOA MAGs from the current study were in agreement with previous findings in that the composition and organization of A-type ATPase gene operons were like those in other AOA isolates from the estuarine and coastal group.
Nitric oxide (NO) is a central intermediate of archaeal ammonia oxidation. A putative copper-containing nitrite reductase (encoded by nirK) may be responsible for NO production and is widely encoded by marine and soil AOA species (69, 70). A single-copy nirK gene was identified in the current study, which was consistent with observations for other marine AOA species from the family Nitrosopumilaceae. No urea utilization gene (ureC) was detected in the AOA MAGs from the Jiulong River estuary. Since the concentration of ammonia is relatively high in the estuary because of river runoff, estuary AOA might not need to use urea as a nitrogen source, unlike in the pelagic ocean.
Flagellation is widespread in archaea and is important for cell motility, DNA uptake, and pathogenicity (71, 72). Most marine Nitrosopumilus strains identified to date (e.g., strains SCM1 [2], PS0 and HCA1 [20], and D3C [73]), except for some cells of strain NF5 (73), lack flagella. However, strains SFB1 and BG20 (genus “Ca. Nitrosoarchaeum”), isolated from estuarine sediments, have flagella and carry the associated genes (25, 74). Further, genes related to the archaeal flagellin were identified in the genome of “Ca. Nitrosopumilus salaria” BD31 in the San Francisco Bay estuary (24). AOA identified in the current study may also have archaeal flagella, as they carry archaeal flagellin genes (flaFGHIJ) and the preflagellin peptidase gene (flaK).
Chemotaxis is an important characteristic of estuarine AOA species, which harbor the related proteins (encoded by cheB) and response regulators (encoded by cheY) (25, 74). These genes were also identified in the novel AOA in the current study. Further, mechanosensitive channel proteins can protect AOA against osmotic stress in the estuary, and the encoding genes were also identified here. Although the relationship between archaeal flagella and AOA ecotype distribution is not well understood, AOA may thrive in the dynamic estuarine environment by relying on cell mobility.
CAZys are a prerequisite for cellular metabolism in diverse carbohydrates, such as glycoproteins, glycolipids, and polysaccharides (75). Chemoautotrophic AOA are currently considered to be mixotrophic in certain environments, which is supported by the presence of CAZy-encoding genes and extracellular peptidases detected in AOA genomes in pure cultures (20) and pelagic (76) and coastal (36) environments. Genes for several glycosyltransferases from the families GT2, GT1, and GT66 were identified in the novel AOA, and also genes for extracellular peptidase (gamma-glutamyltranspeptidase) and serine protease. Notably, the novel AOA genus uniquely harbors chitinase (GH18) genes, which are lacking in other AOA isolates from the family Nitrosopumilaceae. Eutrophication-induced hypoxia and algal blooms are often reported in the Jiulong River estuary (27, 77), and hence, chitin is one of the most abundant carbon sources derived from dead organisms in the local ecosystem. Chitinases are common in bacteria and eukaryotes, and a possible horizontal transfer of chitinase genes may imply potentially close interactions between AOA and other microorganisms. Production of diverse CAZys and extracellular peptidases may expand the capacity for carbohydrate metabolism in AOA and be advantageous in the presence of a variety of anthropogenic pollutants, including organic fertilizers, pesticides, and polycyclic aromatic hydrocarbons, in the Jiulong River estuary.
Massive anthropogenic input influences the local ecosystems in the Jiulong River estuary, resulting in eutrophication and predominantly phosphorus-limited conditions (77–79). Therefore, the acquisition of phosphorus is extremely important for organisms if they have to survive in and adapt to that environment. A recent study suggested that AOA uniquely encode high-affinity phosphate transporters and inorganic-phosphate-dependent regulators to adapt to the phosphorus limitation in the Pearl River estuary (36). No genes encoding high-affinity phosphate transporters were identified in the AOA MAGs in the current study. However, the MAGs encode phosphonate transporters and additional phosphatases, which might constitute a strategy for acquiring phosphorus from diverse substrates. Further, they harbor genes for polyphosphate utilization (ppA), which may be essential for utilizing intracellular phosphate reserves in response to phosphorus starvation (80–82).
Conclusions.
In summary, we have presented potential explanations for the shift of ammonia oxidizers along the Jiulong River estuary and have clearly demonstrated the abundance, diversity, and distribution of AOA in the region. Further, we reconstructed five high-quality MAGs belonging to a novel potential estuarine AOA genus, “Ca. Nitrosopumilaceae” nov. gen., and characterized their genomic features. Genomic comparisons underlined the unique genomic characteristics of estuarine AOA that enable them to thrive in the local environment. Further, pangenomic analysis revealed the potential survival strategies of AOA dwelling in the eutrophic and polluted Jiulong River estuary. The genomic information for estuarine AOA from different ecological niches may further enhance our understanding of the evolution and adaptation of the AOA community and highlights their importance in global nitrification.
MATERIALS AND METHODS
Sample collection, DNA extraction, and sequencing.
Three water samples and three sediment samples were collected in December 2018 during a cruise on the Jiulong River estuary (Fig. 1A). At each sampling station, 500 ml of water was collected using a conductivity-temperature-depth rosette system (CTD) (General Oceanics) in X-Niskin bottles (General Oceanics) 2 m below the surface. The water was then filtered through 0.22-μm polycarbonate filter (EMD Millipore). The surface sediment was collected using a grabber and immediately sealed in 50-ml tubes (Falcon). The filters and sediments were quickly frozen in liquid nitrogen and stored at −80°C until DNA extraction was performed. In situ environmental parameters (depth, salinity, and pH) were measured by using the CTD. The DO concentration was determined using the Winkler method (83). The concentrations of nutrients (NO3−, NO2−, NH4+, and PO43−) were determined using an autoanalyzer (QuAAtro; BLTEC Co., Ltd.). Chlorophyll (Chl) a was processed following the Joint Global Ocean Flux Study protocol (84). Microbial nitrification rates were measured using a dark isotopic incubation approach (85). DNA was extracted using a PowerMax soil kit (Qiagen), following the manufacturer’s instructions. The DNA fragments were purified and then end repaired, A tailed, and ligated with Illumina-compatible adapters before sequencing, according to the method in a previous study (36). Shotgun sequencing was performed using the Illumina (San Diego, CA, USA) HiSeq 2000 paired-end 150-bp platform with a HiSeq cluster kit v4.
Quantitative-PCR analysis of the microbial community.
The determination of the copy numbers of the 16S rRNA gene for all prokaryotes and total archaea and the amoA gene for bacteria and archaea was performed via qPCR using a QuantStudio 3 instrument (Thermo Fisher Scientific). Primer pairs and settings were based on previous reports, as follows: Uni515F/Uni806R for total prokaryotic 16S rRNA genes (86), Arch519F/Arch908R for total archaeal 16S rRNA genes (87, 88), amoA-1F/amoA-2R for bacterial amoA genes (89, 90), and crenamoA23f/crenamoA616r for archaeal amoA genes (6, 8) (Table 3; see Table S1). The analyses were done in triplicate for each sample and were performed in 20-μl qPCR mixtures containing 10 μl of PowerUp SYBR green master mix (Applied Biosystems), 2 μl of DNA template, 0.5 μl of each of the forward and reverse primers (10 μM), and 7 μl of double-distilled water (ddH2O). Standard qPCR curves were generated using a sequential 10-fold dilution series of the pMD19-T vector, as previously described (91). Gel electrophoretograms for each primer pair and melting curves for each qPCR experiment were used to verify the results. Gene copy numbers in the standard dilution series were calculated by first determining the DNA concentration with a Nanodrop (Thermo Scientific) and then using the following equation: gene copies per microliter = (amount per microliter × 6.022 × 1023)/(length × 1 × 109 × 324.5).
TABLE 3.
Primers and conditions used for qPCR experiments
| Target gene | Target description | Primer name | Sequence (5′-3′) | Annealing temp (°C) | R2 | Efficiency (%) | Reference(s) |
|---|---|---|---|---|---|---|---|
| 16S rRNA | Bacteria plus archaea | Uni515F | GTGYCAGCMGCCGCGGTAA | 60 | 0.998 | 87.63 | 86 |
| Uni806R | GGACTACNVGGGTWTCTAAT | ||||||
| 16S rRNA | Archaea | Arch519F | CAGCCGCCGCGGTAA | 60 | 0.994 | 85.17 | 87, 88 |
| Arch908R | CCCGCC AATTCCTTT AAGTT | ||||||
| amoA | AOB amoA genes | amoA-1F | GGGGHTTYTACTGGTGGT | 55 | 0.993 | 101 | 89, 90 |
| amoA-2R | CCCCTCKGSAAAGCCTTCTTC | ||||||
| amoA | AOA amoA genes | CrenamoA23f | ATGGTCTGGCTWAGACG | 55 | 0.997 | 98.8 | 6, 8 |
| CrenamoA616r | GCCATCCABCKRTANGTCCA |
Metagenomic assembly, genome binning, and annotation.
Raw reads after metagenomic shotgun sequencing were dereplicated at 100% identity and trimmed using Sickle (https://github.com/najoshi/sickle) with default settings. The trimmed forward and reverse reads from each sample were de novo assembled using IDBA-UD (version 1.1.1) with the following parameters: -mink 65, -maxk 145, and -step 10 (92). The small-subunit (SSU) rRNA genes were extracted from the assembled scaffolds for each sample using Metaxa2 software (version 2.2) (93). For each sample, Metabat2 (version 2.12.1) was employed for automatic binning from the assembly 12 times with different parameters for further refinement (94). All the binning results were merged and then refined using the standard pipeline of DASTools (version 1.0) (95). In addition, the genomes were reassembled and refined under the standard procedures of anvio (version 5.5) for improved data quality before downstream analysis (96). MAG completeness, contamination, and strain heterogeneity were then determined by using CheckM (97). MAGs with completeness exceeding 60% and contamination below 10% were selected for further analysis.
rRNA coding regions (16S and 23S rRNA) in each genome were identified using both CheckM SSU_finder (97) and Metaxa2 (93). Genes were called by Prodigal (version 2.6.3) with the “−p meta” option for each genome (98). The genes were annotated using the KEGG server (BlastKOALA) (99) and BLASTP against the nonredundant protein database (October 2016 version; E value cutoff, <1e−5). Further, all proteins were assigned to existing COG and archaeal clusters of orthologous genes (arCOG) by eggNOG-mapper (100). PRED-SIGNAL (101) and PSORTb (102) were used to identify extracellular peptidases, and the dbCAN Web server (103) was used for carbohydrate-active-gene identification.
Phylogenetic analysis.
The amoA gene sequences extracted from the MAGs and assembled scaffolds were aligned using Mafft-LINSi (104) and categorized with reference sequences, as previously described (16). The maximum-likelihood phylogenetic tree of amoA genes was inferred using IQ-TREE (105) and the best-fit mtZOA+G4 model selected by the -m TEST option with ultrafast bootstrapping (-bb 1,000) (106). The 16S and 23S rRNA genes of MAGs and references (16) were aligned and concatenated by Mafft (104). The maximum-likelihood phylogeny tree of concatenated 16S and 23S rRNA genes was inferred using IQ-TREE and a GTR+F+R6 model with ultrafast bootstrapping (-bb 1,000). The maximum-likelihood phylogenetic tree of MAGs was constructed using the IQ-TREE (LG+C60+F model), based on 15 concatenated ribosomal proteins (L2, L3, L4, L5, L6, L14, L15e, L18p, L22, L24e, S3Ae, S8, S11, S17, and S19) extracted from MAGs from the current study and some major archaeal phyla (reference genomes) using the anvio 5 pipeline. Chitinase protein sequences extracted from AOA MAGs from the current study were aligned with other bacterial and archaeal chitinase reference sequences using Mafft (104); the phylogenetic tree of chitinase proteins was built using IQ-TREE. All the phylogenetic trees were visualized using the online tool iTOL (version 5.0) (107). To indicate the abundances of amoA genes and 16S rRNA genes, the number of reads per kilobase per million sequenced reads (RPKM) was used to normalize the variations in gene length and data set size. The relative abundances for MAGs in each sample were normalized with the following formula, as described previously (36): MAG coverage/total coverage for each sample.
Genomic comparisons and analysis.
The AAI between MAGs from the current study and genomes of AOA representing different archaeal genera or species was calculated using compareM (https://github.com/dparks1134/CompareM). The similarity of 16S rRNA genes of MAGs from the current study and reference AOA genomes was determined by pairwise BLASTN (E value cutoff, <1e−5). AOA MAGs from the current study and other reported genomes, such as those of “Ca. Nitrosopumilus salaria” BD31 (24) and “Ca. Nitrosoarchaeum” strains SFB1 and BG20 (25), and MAGs (36) within the family Nitrosopumilaceae obtained from estuaries were combined in an estuarine gene pool and compared with the marine AOA gene pool (i.e., strains SCM1, D3C, NF5, AR1, AR2, and SPOT01) based on COG functional categories. Similarly, MAGs of the novel AOA species identified in the current study were pooled in a single gene pool to identify composition differences and unique genes by comparison with other AOA species within the genus Nitrosopumilus. Pangenomic analysis of MAGs from the current study and AOA isolates/enrichments from the genus Nitrosopumilus was performed using the anvio 5 pangenome pipeline (108). Genes unique to the genomes reported in the current study were identified and then annotated by BLASTP analysis against the nonredundant protein database (October 2016 version; E value cutoff, <1e−5).
Data availability.
The raw reads for all six samples are available in the National Omics Data Encyclopedia (NODE) database (http://www.biosino.org/node) under accession numbers OER057292 to OER057297, and nine AOA genomic bins are available under accession numbers OER057298 to OER057306.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the National Natural Science Foundation of China (grant no. 91851105, 31970105, and 31622002), the Shenzhen Science and Technology Program (grant no. JCYJ20170818091727570 and KQTD20180412181334790), the Key Project of Department of Education of Guangdong Province (grant no. 2017KZDXM071), and the CAS Interdisciplinary Innovation Team (grant no. JCTD-2018-16).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Kowalchuk GA, Stephen JR. 2001. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu Rev Microbiol 55:485–529. doi: 10.1146/annurev.micro.55.1.485. [DOI] [PubMed] [Google Scholar]
- 2.Könneke M, Bernhard AE, José R, Walker CB, Waterbury JB, Stahl DA. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 3.Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A 102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJM, Kartal B, Jetten MSM, Lücker S. 2015. Complete nitrification by a single microorganism. Nature 528:555–559. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M. 2015. Complete nitrification by Nitrospira bacteria. Nature 528:504–509. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster S, Schleper C. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- 7.Nunoura T, Takaki Y, Hirai M, Shimamura S, Makabe A, Koide O, Kikuchi T, Miyazaki J, Koba K, Yoshida N, Sunamura M, Takai K. 2015. Hadal biosphere: insight into the microbial ecosystem in the deepest ocean on Earth. Proc Natl Acad Sci U S A 112:E1230–E1236. doi: 10.1073/pnas.1421816112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicol GW, Leininger S, Schleper C, Prosser JI. 2008. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol 10:2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- 9.Reigstad LJ, Richter A, Daims H, Urich T, Schwark L, Schleper C. 2008. Nitrification in terrestrial hot springs of Iceland and Kamchatka. FEMS Microbiol Ecol 64:167–174. doi: 10.1111/j.1574-6941.2008.00466.x. [DOI] [PubMed] [Google Scholar]
- 10.Pester M, Schleper C, Wagner M. 2011. The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol 14:300–306. doi: 10.1016/j.mib.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sintes E, De Corte D, Haberleitner E, Herndl GJ. 2016. Geographic distribution of archaeal ammonia oxidizing ecotypes in the Atlantic Ocean. Front Microbiol 7:77. doi: 10.3389/fmicb.2016.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B, Qin W, Ren Y, Zhou X, Jung M-Y, Han P, Eloe-Fadrosh EA, Li M, Zheng Y, Lu L, Yan X, Ji J, Liu Y, Liu L, Heiner C, Hall R, Martens-Habbena W, Herbold CW, Rhee S-K, Bartlett DH, Huang L, Ingalls AE, Wagner M, Stahl DA, Jia Z. 2019. Expansion of Thaumarchaeota habitat range is correlated with horizontal transfer of ATPase operons. ISME J 13:3067–3013. doi: 10.1038/s41396-019-0493-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stieglmeier M, Alves R, Schleper C. 2014. Thaumarchaeota, p 347–362. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes: other major lineages of Bacteria and the Archaea. Springer, Berlin, Germany. [Google Scholar]
- 14.Smith JM, Casciotti KL, Chavez FP, Francis CA. 2014. Differential contributions of archaeal ammonia oxidizer ecotypes to nitrification in coastal surface waters. ISME J 8:1704–1714. doi: 10.1038/ismej.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung S, Mak W, Xia X, Lu Y, Cheung Y, Liu H. 2019. Overlooked genetic diversity of ammonia oxidizing archaea lineages in the global oceans. J Geophys Res Biogeosci 124:1799–1811. doi: 10.1029/2018JG004636. [DOI] [Google Scholar]
- 16.Alves RJE, Minh BQ, Urich T, von Haeseler A, Schleper C. 2018. Unifying the global phylogeny and environmental distribution of ammonia-oxidising archaea based on amoA genes. Nat Commun 9:1517. doi: 10.1038/s41467-018-03861-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahlgren NA, Chen Y, Needham DM, Parada AE, Sachdeva R, Trinh V, Chen T, Fuhrman JA. 2017. Genome and epigenome of a novel marine Thaumarchaeota strain suggest viral infection, phosphorothioation DNA modification and multiple restriction systems. Environ Microbiol 19:2434–2452. doi: 10.1111/1462-2920.13768. [DOI] [PubMed] [Google Scholar]
- 18.Jung M-Y, Kim J-G, Sinninghe Damsté JS, Rijpstra WIC, Madsen EL, Kim S-J, Hong H, Si O-J, Kerou M, Schleper C, Rhee S-K. 2016. A hydrophobic ammonia‐oxidizing archaeon of the Nitrosocosmicus clade isolated from coal tar‐contaminated sediment. Environ Microbiol Rep 8:983–992. doi: 10.1111/1758-2229.12477. [DOI] [PubMed] [Google Scholar]
- 19.Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C. 2011. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci U S A 108:8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin W, Amin SA, Martens-Habbena W, Walker CB, Urakawa H, Devol AH, Ingalls AE, Moffett JW, Armbrust EV, Stahl DA. 2014. Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc Natl Acad Sci U S A 111:12504–12509. doi: 10.1073/pnas.1324115111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lehtovirta-Morley LE, Ge C, Ross J, Yao H, Nicol GW, Prosser JI. 2014. Characterisation of terrestrial acidophilic archaeal ammonia oxidisers and their inhibition and stimulation by organic compounds. FEMS Microbiol Ecol 89:542–552. doi: 10.1111/1574-6941.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daebeler A, Herbold CW, Vierheilig J, Sedlacek CJ, Pjevac P, Albertsen M, Kirkegaard RH, De La Torre JR, Daims H, Wagner M. 2018. Cultivation and genomic analysis of “Candidatus Nitrosocaldus islandicus,” an obligately thermophilic, ammonia-oxidizing thaumarchaeon from a hot spring biofilm in Graendalur Valley, Iceland. Front Microbiol 9:193. doi: 10.3389/fmicb.2018.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abby SS, Melcher M, Kerou M, Krupovic M, Stieglmeier M, Rossel C, Pfeifer K, Schleper C. 2018. Candidatus Nitrosocaldus cavascurensis, an ammonia oxidizing, extremely thermophilic archaeon with a highly mobile genome. Front Microbiol 9:28. doi: 10.3389/fmicb.2018.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mosier AC, Allen EE, Kim M, Ferriera S, Francis CA. 2012. Genome sequence of “Candidatus Nitrosopumilus salaria” BD31, an ammonia-oxidizing archaeon from the San Francisco Bay estuary. J Bacteriol 194:2121–2122. doi: 10.1128/JB.00013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mosier AC, Lund MB, Francis CA. 2012. Ecophysiology of an ammonia-oxidizing archaeon adapted to low-salinity habitats. Microb Ecol 64:955–963. doi: 10.1007/s00248-012-0075-1. [DOI] [PubMed] [Google Scholar]
- 26.Luo Z, Qiu Z, Wei Q, Du Laing G, Zhao Y, Yan C. 2014. Dynamics of ammonia-oxidizing archaea and bacteria in relation to nitrification along simulated dissolved oxygen gradient in sediment-water interface of the Jiulong River estuarine wetland, China. Environ Earth Sci 72:2225–2237. doi: 10.1007/s12665-014-3128-6. [DOI] [Google Scholar]
- 27.Wu J, Chen N, Hong H, Lu T, Wang L, Chen Z. 2013. Direct measurement of dissolved N2 and denitrification along a subtropical river-estuary gradient, China. Mar Pollut Bull 66:125–134. doi: 10.1016/j.marpolbul.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 28.Li Q, Wang F, Chen Z, Yin X, Xiao X. 2012. Stratified active archaeal communities in the sediments of Jiulong River estuary, China. Front Microbiol 3:311. doi: 10.3389/fmicb.2012.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu A, Hou L, Yu C-P. 2015. Biogeography of planktonic and benthic archaeal communities in a subtropical eutrophic estuary of China. Microb Ecol 70:322–335. doi: 10.1007/s00248-015-0597-4. [DOI] [PubMed] [Google Scholar]
- 30.Murray AE, Freudenstein J, Gribaldo S, Hatzenpichler R, Hugenholtz P, Kämpfer P, Konstantinidis KT, Lane CE, Papke RT, Parks DH. 2020. Roadmap for naming uncultivated Archaea and Bacteria. Nat Microbiol 5:987–994. doi: 10.1038/s41564-020-0733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konstantinidis KT, Tiedje JM. 2005. Towards a genome-based taxonomy for prokaryotes. J Bacteriol 187:6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konstantinidis KT, Tiedje JM. 2007. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr Opin Microbiol 10:504–509. doi: 10.1016/j.mib.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 33.Offre P, Kerou M, Spang A, Schleper C. 2014. Variability of the transporter gene complement in ammonia-oxidizing archaea. Trends Microbiol 22:665–675. doi: 10.1016/j.tim.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Shiozaki T, Ijichi M, Isobe K, Hashihama F, Nakamura K-i, Ehama M, Hayashizaki K-I, Takahashi K, Hamasaki K, Furuya K. 2016. Nitrification and its influence on biogeochemical cycles from the equatorial Pacific to the Arctic Ocean. ISME J 10:2184–2197. doi: 10.1038/ismej.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clark DR, Rees AP, Joint I. 2008. Ammonium regeneration and nitrification rates in the oligotrophic Atlantic Ocean: implications for new production estimates. Limnol Oceanogr 53:52–62. doi: 10.4319/lo.2008.53.1.0052. [DOI] [Google Scholar]
- 36.Zou D, Li Y, Kao SJ, Liu H, Li M. 2019. Genomic adaptation to eutrophication of ammonia‐oxidizing archaea in the Pearl River estuary. Environ Microbiol 21:2320–2332. doi: 10.1111/1462-2920.14613. [DOI] [PubMed] [Google Scholar]
- 37.Wang W, Yu Z, Wu Z, Song S, Song X, Yuan Y, Cao X. 2018. Rates of nitrification and nitrate assimilation in the Changjiang River estuary and adjacent waters based on the nitrogen isotope dilution method. Continental Shelf Res 163:35–43. doi: 10.1016/j.csr.2018.04.014. [DOI] [Google Scholar]
- 38.Damashek J, Casciotti KL, Francis CA. 2016. Variable nitrification rates across environmental gradients in turbid, nutrient-rich estuary waters of San Francisco Bay. Estuaries Coasts 39:1050–1071. doi: 10.1007/s12237-016-0071-7. [DOI] [Google Scholar]
- 39.Vipindas P, Anas A, Jayalakshmy K, Lallu K, Benny P, Shanta N. 2018. Impact of seasonal changes in nutrient loading on distribution and activity of nitrifiers in a tropical estuary. Continental Shelf Res 154:37–45. doi: 10.1016/j.csr.2018.01.003. [DOI] [Google Scholar]
- 40.Sanders T, Laanbroek HJ. 2018. The distribution of sediment and water column nitrification potential in the hyper-turbid Ems estuary. Aquat Sci 80:33. doi: 10.1007/s00027-018-0584-1. [DOI] [Google Scholar]
- 41.Kuypers MM, Marchant HK, Kartal B. 2018. The microbial nitrogen-cycling network. Nat Rev Microbiol 16:263–276. doi: 10.1038/nrmicro.2018.9. [DOI] [PubMed] [Google Scholar]
- 42.Xia F, Wang J-G, Zhu T, Zou B, Rhee S-K, Quan Z-X. 2018. Ubiquity and diversity of complete ammonia oxidizers (comammox). Appl Environ Microbiol 84:e01390-18. doi: 10.1128/AEM.01390-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koch H, van Kessel MA, Lücker S. 2019. Complete nitrification: insights into the ecophysiology of comammox Nitrospira. Appl Microbiol Biotechnol 103:177–189. doi: 10.1007/s00253-018-9486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Ma L, Mao Y, Jiang X, Xia Y, Yu K, Li B, Zhang T. 2017. Comammox in drinking water systems. Water Res 116:332–341. doi: 10.1016/j.watres.2017.03.042. [DOI] [PubMed] [Google Scholar]
- 45.Liu Z, Huang S, Sun G, Xu Z, Xu M. 2011. Diversity and abundance of ammonia-oxidizing archaea in the Dongjiang River, China. Microbiol Res 166:337–345. doi: 10.1016/j.micres.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 46.Li Z, Jin W, Liang Z, Yue Y, Lv J. 2013. Abundance and diversity of ammonia-oxidizing archaea in response to various habitats in Pearl River Delta of China, a subtropical maritime zone. J Environ Sci 25:1195–1205. doi: 10.1016/S1001-0742(12)60178-8. [DOI] [PubMed] [Google Scholar]
- 47.Cao H, Hong Y, Li M, Gu J-D. 2011. Diversity and abundance of ammonia-oxidizing prokaryotes in sediments from the coastal Pearl River estuary to the South China Sea. Antonie Van Leeuwenhoek 100:545–556. doi: 10.1007/s10482-011-9610-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mosier AC, Francis CA. 2008. Relative abundance and diversity of ammonia‐oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ Microbiol 10:3002–3016. doi: 10.1111/j.1462-2920.2008.01764.x. [DOI] [PubMed] [Google Scholar]
- 49.Bouvier TC, del Giorgio PA. 2002. Compositional changes in free‐living bacterial communities along a salinity gradient in two temperate estuaries. Limnol Oceanogr 47:453–470. doi: 10.4319/lo.2002.47.2.0453. [DOI] [Google Scholar]
- 50.Campbell BJ, Kirchman DL. 2013. Bacterial diversity, community structure and potential growth rates along an estuarine salinity gradient. ISME J 7:210–220. doi: 10.1038/ismej.2012.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Norton JM. 2011. Diversity and environmental distribution of ammonia-oxidizing bacteria, p 39–55. In Ward BB, Klotz MG, Arp DJ (ed), Nitrification. American Society for Microbiology, Washington, DC. [Google Scholar]
- 52.Speksnijder AG, Kowalchuk GA, Roest K, Laanbroek HJ. 1998. Recovery of a Nitrosomonas-like 16S rDNA sequence group from freshwater habitats. Syst Appl Microbiol 21:321–330. doi: 10.1016/S0723-2020(98)80040-4. [DOI] [PubMed] [Google Scholar]
- 53.Belser L, Schmidt E. 1978. Diversity in the ammonia-oxidizing nitrifier population of a soil. Appl Environ Microbiol 36:584–588. doi: 10.1128/AEM.36.4.584-588.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dang H, Li J, Chen R, Wang L, Guo L, Zhang Z, Klotz MG. 2010. Diversity, abundance, and spatial distribution of sediment ammonia-oxidizing Betaproteobacteria in response to environmental gradients and coastal eutrophication in Jiaozhou Bay, China. Appl Environ Microbiol 76:4691–4702. doi: 10.1128/AEM.02563-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Regan JM, Harrington GW, Noguera DR. 2002. Ammonia-and nitrite-oxidizing bacterial communities in a pilot-scale chloraminated drinking water distribution system. Appl Environ Microbiol 68:73–81. doi: 10.1128/aem.68.1.73-81.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao J, Luo X, Wu G, Li T, Peng Y. 2014. Abundance and diversity based on amoA genes of ammonia-oxidizing archaea and bacteria in ten wastewater treatment systems. Appl Microbiol Biotechnol 98:3339–3354. doi: 10.1007/s00253-013-5428-2. [DOI] [PubMed] [Google Scholar]
- 57.Stehr G, Böttcher B, Dittberner P, Rath G, Koops H-P. 1995. The ammonia-oxidizing nitrifying population of the River Elbe estuary. FEMS Microbiol Ecol 17:177–186. doi: 10.1111/j.1574-6941.1995.tb00141.x. [DOI] [Google Scholar]
- 58.Rysgaard S, Thastum P, Dalsgaard T, Christensen PB, Sloth NP, Rysgaard S. 1999. Effects of salinity on NH 4+ adsorption capacity, nitrification, and denitrification in Danish estuarine sediments. Estuaries 22:21–30. doi: 10.2307/1352923. [DOI] [Google Scholar]
- 59.Bernhard AE, Landry ZC, Blevins A, José R, Giblin AE, Stahl DA. 2010. Abundance of ammonia-oxidizing archaea and bacteria along an estuarine salinity gradient in relation to potential nitrification rates. Appl Environ Microbiol 76:1285–1289. doi: 10.1128/AEM.02018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Chen L, Dai T, Tian J, Wen D. 2015. The influence of salinity on the abundance, transcriptional activity, and diversity of AOA and AOB in an estuarine sediment: a microcosm study. Appl Microbiol Biotechnol 99:9825–9833. doi: 10.1007/s00253-015-6804-x. [DOI] [PubMed] [Google Scholar]
- 61.Martens-Habbena W, Berube PM, Urakawa H, José R, Stahl DA. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 62.Li M, Cao H, Hong Y, Gu J-D. 2011. Spatial distribution and abundances of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in mangrove sediments. Appl Microbiol Biotechnol 89:1243–1254. doi: 10.1007/s00253-010-2929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cao H, Li M, Hong Y, Gu J-D. 2011. Diversity and abundance of ammonia-oxidizing archaea and bacteria in polluted mangrove sediment. Syst Appl Microbiol 34:513–523. doi: 10.1016/j.syapm.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 64.Zheng Y, Hou L, Liu M, Lu M, Zhao H, Yin G, Zhou J. 2013. Diversity, abundance, and activity of ammonia-oxidizing bacteria and archaea in Chongming eastern intertidal sediments. Appl Microbiol Biotechnol 97:8351–8363. doi: 10.1007/s00253-012-4512-3. [DOI] [PubMed] [Google Scholar]
- 65.Sun W, Xia C, Xu M, Guo J, Wang A, Sun G. 2013. Distribution and abundance of archaeal and bacterial ammonia oxidizers in the sediments of the Dongjiang River, a drinking water supply for Hong Kong. Microb Environ 28:457–465. doi: 10.1264/jsme2.ME13066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dang H, Zhou H, Yang J, Ge H, Jiao N, Luan X, Zhang C, Klotz MG. 2013. Thaumarchaeotal signature gene distribution in sediments of the northern South China Sea: an indicator of the metabolic intersection of the marine carbon, nitrogen, and phosphorus cycles? Appl Environ Microbiol 79:2137–2147. doi: 10.1128/AEM.03204-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Herbold CW, Lehtovirta-Morley LE, Jung M-Y, Jehmlich N, Hausmann B, Han P, Loy A, Pester M, Sayavedra-Soto LA, Rhee S-K, Prosser JI, Nicol GW, Wagner M, Gubry-Rangin C. 2017. Ammonia‐oxidising archaea living at low pH: insights from comparative genomics. Environ Microbiol 19:4939–4952. doi: 10.1111/1462-2920.13971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Könneke M, Schubert DM, Brown PC, Hügler M, Standfest S, Schwander T, von Borzyskowski LS, Erb TJ, Stahl DA, Berg IA. 2014. Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation. Proc Natl Acad Sci U S A 111:8239–8244. doi: 10.1073/pnas.1402028111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carini P, Dupont CL, Santoro AE. 2018. Patterns of thaumarchaeal gene expression in culture and diverse marine environments. Environ Microbiol 20:2112–2124. doi: 10.1111/1462-2920.14107. [DOI] [PubMed] [Google Scholar]
- 70.Qin W, Amin SA, Lundeen RA, Heal KR, Martens-Habbena W, Turkarslan S, Urakawa H, Costa KC, Hendrickson EL, Wang T, Beck DA, Tiquia-Arashiro SM, Taub F, Holmes AD, Vajrala N, Berube PM, Lowe TM, Moffett JW, Devol AH, Baliga NS, Arp DJ, Sayavedra-Soto LA, Hackett M, Armbrust EV, Ingalls AE, Stahl DA. 2018. Stress response of a marine ammonia-oxidizing archaeon informs physiological status of environmental populations. ISME J 12:508–519. doi: 10.1038/ismej.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jarrell KF, Bayley DP, Kostyukova AS. 1996. The archaeal flagellum: a unique motility structure. J Bacteriol 178:5057–5064. doi: 10.1128/jb.178.17.5057-5064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Albers S-V, Meyer BH. 2011. The archaeal cell envelope. Nat Rev Microbiol 9:414–426. doi: 10.1038/nrmicro2576. [DOI] [PubMed] [Google Scholar]
- 73.Bayer B, Vojvoda J, Offre P, Alves RJ, Elisabeth NH, Garcia JA, Volland J-M, Srivastava A, Schleper C, Herndl GJ. 2016. Physiological and genomic characterization of two novel marine thaumarchaeal strains indicates niche differentiation. ISME J 10:1051–1063. doi: 10.1038/ismej.2015.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. 2011. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One 6:e16626. doi: 10.1371/journal.pone.0016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Davies GJ, Gloster TM, Henrissat B. 2005. Recent structural insights into the expanding world of carbohydrate-active enzymes. Curr Opin Struct Biol 15:637–645. doi: 10.1016/j.sbi.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 76.Li M, Baker BJ, Anantharaman K, Jain S, Breier JA, Dick GJ. 2015. Genomic and transcriptomic evidence for scavenging of diverse organic compounds by widespread deep-sea archaea. Nat Commun 6:8933. doi: 10.1038/ncomms9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y, Cao W, Su C, Hong H. 2011. Nutrient sources and composition of recent algal blooms and eutrophication in the northern Jiulong River, Southeast China. Mar Pollut Bull 63:249–254. doi: 10.1016/j.marpolbul.2011.02.021. [DOI] [PubMed] [Google Scholar]
- 78.Chen N, Peng B, Hong H, Turyaheebwa N, Cui S, Mo X. 2013. Nutrient enrichment and N:P ratio decline in a coastal bay-river system in southeast China: the need for a dual nutrient (N and P) management strategy. Ocean Coastal Manag 81:7–13. doi: 10.1016/j.ocecoaman.2012.07.013. [DOI] [Google Scholar]
- 79.Wu G, Cao W, Huang Z, Kao C-M, Chang C-T, Chiang P-C, Wang F. 2017. Decadal changes in nutrient fluxes and environmental effects in the Jiulong River estuary. Mar Pollut Bull 124:871–877. doi: 10.1016/j.marpolbul.2017.01.071. [DOI] [PubMed] [Google Scholar]
- 80.Gómez-García MR, Losada M, Serrano A. 2003. Concurrent transcriptional activation of ppa and ppx genes by phosphate deprivation in the cyanobacterium Synechocystis sp. strain PCC 6803. Biochem Biophys Res Commun 302:601–609. doi: 10.1016/s0006-291x(03)00162-1. [DOI] [PubMed] [Google Scholar]
- 81.Peimbert M, Alcaraz LD, Bonilla-Rosso G, Olmedo-Alvarez G, García-Oliva F, Segovia L, Eguiarte LE, Souza V. 2012. Comparative metagenomics of two microbial mats at Cuatro Ciénegas Basin I: ancient lessons on how to cope with an environment under severe nutrient stress. Astrobiology 12:648–658. doi: 10.1089/ast.2011.0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pereira N, Shilova IN, Zehr JP. 2016. Molecular markers define progressing stages of phosphorus limitation in the nitrogen‐fixing cyanobacterium, Crocosphaera. J Phycol 52:274–282. doi: 10.1111/jpy.12396. [DOI] [PubMed] [Google Scholar]
- 83.Hitchman ML. 1978. Measurement of dissolved oxygen. Wiley, New York, NY. [Google Scholar]
- 84.Knap A, Michaels A, Close A, Ducklow H, Dickson A. 1996. Protocols for the Joint Global Ocean Flux Study (JGOFS) core measurements. UNESCO, Paris, France. [Google Scholar]
- 85.Hou L, Xie X, Wan X, Kao S-J, Jiao N, Zhang Y. 2018. Niche differentiation of ammonia and nitrite oxidizers along a salinity gradient from the Pearl River estuary to the South China Sea. Biogeosciences 15:5169–5187. doi: 10.5194/bg-15-5169-2018. [DOI] [Google Scholar]
- 86.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ovreås L, Forney L, Daae FL, Torsvik V. 1997. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol 63:3367–3373. doi: 10.1128/AEM.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jorgensen SL, Hannisdal B, Lanzén A, Baumberger T, Flesland K, Fonseca R, Ovreås L, Steen IH, Thorseth IH, Pedersen RB, Schleper C. 2012. Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc Natl Acad Sci U S A 109:E2846–E2855. doi: 10.1073/pnas.1207574109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stephen JR, Chang Y-J, MacNaughton SJ, Kowalchuk GA, Leung KT, Flemming CA, White DC. 1999. Effect of toxic metals on indigenous soil β-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl Environ Microbiol 65:95–101. doi: 10.1128/AEM.65.1.95-101.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rotthauwe J-H, Witzel K-P, Liesack W. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712. doi: 10.1128/AEM.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pan J, Chen Y, Wang Y, Zhou Z, Li M. 2019. Vertical distribution of bathyarchaeotal communities in mangrove wetlands suggests distinct niche preference of Bathyarchaeota subgroup 6. Microb Ecol 77:417–428. doi: 10.1007/s00248-018-1309-7. [DOI] [PubMed] [Google Scholar]
- 92.Peng Y, Leung HC, Yiu S-M, Chin FY. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 93.Bengtsson‐Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, Larsson DGJ, Nilsson RH. 2015. METAXA2: improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol Ecol Resour 15:1403–1414. doi: 10.1111/1755-0998.12399. [DOI] [PubMed] [Google Scholar]
- 94.Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, Wang Z. 2019. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359. doi: 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sieber CM, Probst AJ, Sharrar A, Thomas BC, Hess M, Tringe SG, Banfield JF. 2018. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol 3:836–843. doi: 10.1038/s41564-018-0171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO. 2015. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 100.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, Von Mering C, Bork P. 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol 34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bagos P, Tsirigos K, Plessas S, Liakopoulos T, Hamodrakas S. 2009. Prediction of signal peptides in archaea. Protein Eng Des Sel 22:27–35. doi: 10.1093/protein/gzn064. [DOI] [PubMed] [Google Scholar]
- 102.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FSL. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. 2012. dbCAN: a Web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Delmont TO, Eren AM. 2018. Linking pangenomes and metagenomes: the Prochlorococcus metapangenome. PeerJ 6:e4320. doi: 10.7717/peerj.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw reads for all six samples are available in the National Omics Data Encyclopedia (NODE) database (http://www.biosino.org/node) under accession numbers OER057292 to OER057297, and nine AOA genomic bins are available under accession numbers OER057298 to OER057306.