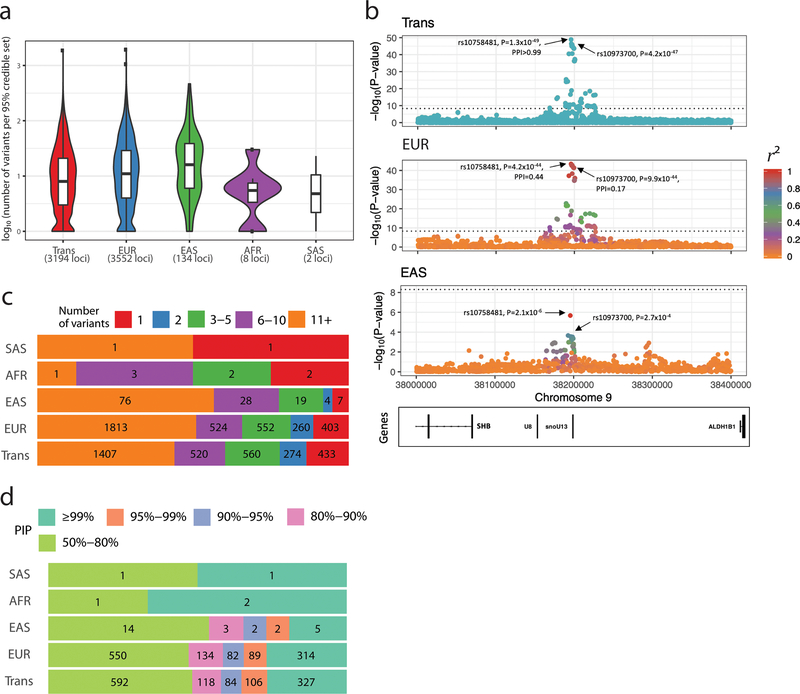
Figure 2.
Fine-mapping of genome-wide significant loci associated with hematological traits. (a) We restricted fine-mapping to loci with evidence for a single association signal in European-ancestry (EUR) populations. There are no such loci in Hispanic Americans. The 95% credible sets in the trans-ethnic meta-analyses are smaller than in the EUR or East-Asian-ancestry (EAS) meta-analyses. (b) Trans-ethnic fine-mapping of a platelet locus. In EUR individuals, the 95% credible set include seven variants with posterior inclusion probability (PIP) >0.04 and strong pairwise linkage disequilibrium (LD) with the sentinel variant rs10758481 (r2>0.93 in British in England and Scotland (GBR) individuals from 1000 Genomes Project, middle panel). LD is similarly strong in African-, Hispanic/South American-, and South-Asian-ancestry populations from the 1000 Genomes Project. However, LD is weaker in East Asians (r2=0.68 in Japanese individuals (JPT) from the 1000 Genomes Project, bottom panel). In the trans-ethnic meta-analysis, rs10758481 has a PIP>0.99 (top panel). In EUR and EAS, LD is color-coded based on pairwise r2 with rs10758481. The dotted line indicates the genome-wide significance threshold (P<5×10−9). ( c) Proportion of 95% credible sets in each population with a defined number of variants. For instance, in the EUR and trans meta-analysis results, we identified 403 and 433 95% credible sets that contain a single variant, respectively. (d) Prioritization of causal variants using fine-mapping PIP. In each population, we provide the proportion of variants with a PIP within a specified range. For instance, in EUR and trans, we found 314 and 327 variants with a PIP ≥99%, respectively. See also Figure S2.