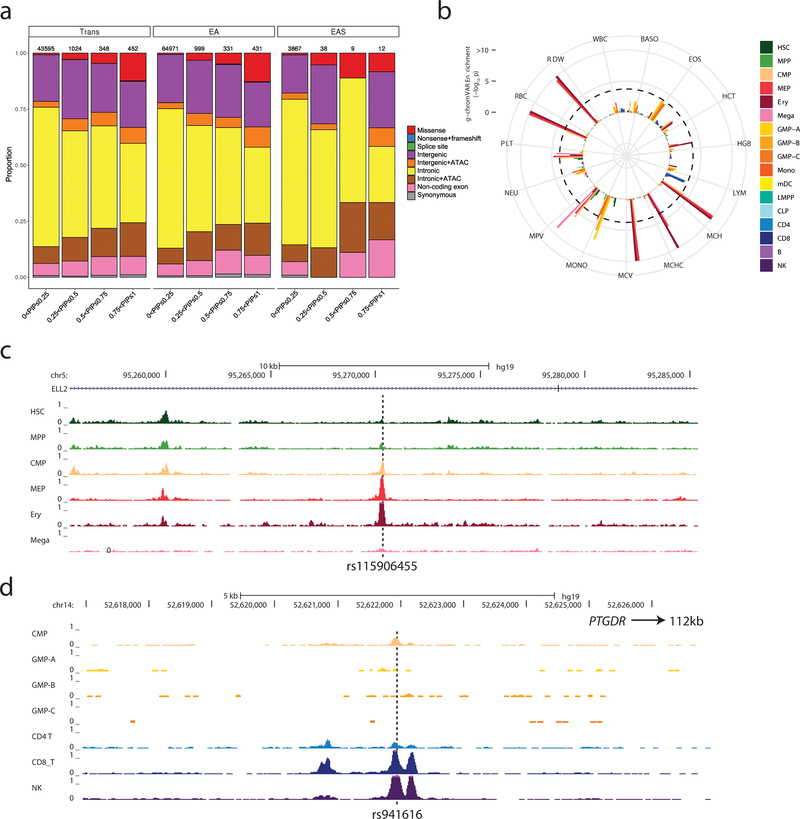
Figure 3.
Functional annotation of possible causal variants associated with blood-cell traits. (a) Annotation of variants in trans, EUR and EAS shows a similar pattern, with a larger proportion of likely functional variants (e.g. missense, intergenic and intronic variants within ATAC-seq peaks) among variants with higher posterior inclusion probability (PIP). (b) g-chromVAR results for trans variants within 95% credible sets for 15 traits. The Bonferroni-adjusted significance level (corrected for 15 traits and 18 cell types) is indicated by the dotted line. Mono, monocyte; HSC, hematopoietic stem cell; Ery, erythroid; Mega, megakaryocyte; CD4, CD4+ T lymphocyte; CD8, CD8+ T lymphocyte; B, B lymphocyte; NK, natural killer cell; mDC, Myeloid dendritic cell; pDC, Plasmacytoid dendritic cell; MPP, multipotent progenitor; LMPP, lymphoid-primed multipotent progenitor; CMP, common myeloid progenitor; CLP, common lymphoid progenitor; GMP, granulocyte–macrophage progenitor; MEP, megakaryocyte–erythroid progenitor. (c) rs115906455 is a novel variant associated with mean corpuscular volume in the trans-ethnic meta-analysis (P=4.2×10−12, PIP=0.57). It maps to an intron of ELL2 and overlaps with ATAC-seq peaks found in CMP, MEP, erythroblasts but not megakaryocytes. (d) rs941616 is a novel variant associated with eosinophil counts in the trans-ethnic meta-analysis (P=2.4×10−9, PIP=0.2). It is a strong eQTL for PTGDR located 112-kb downstream and overlaps with ATAC-seq peaks found in CMP, CD8+ lymphocytes and NK cells. See also Figures S3–4 and Table S3H–I.