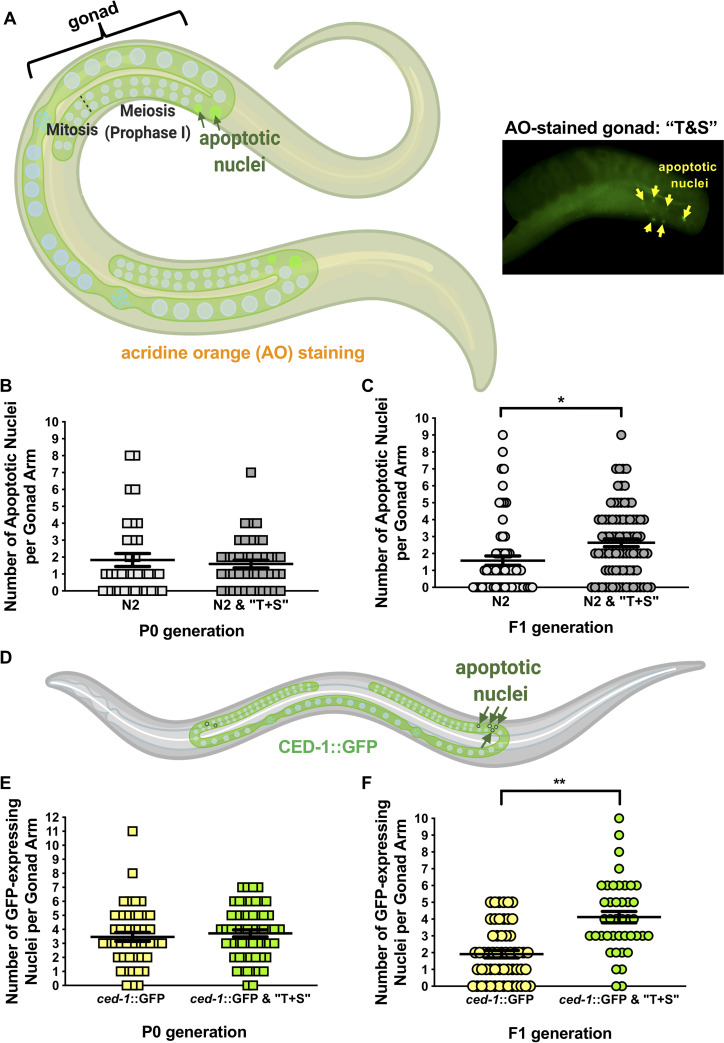
Fig 6. “T+S” exposure results in elevated germline apoptosis in the offspring of wild-type germ lines.
Wild-type germ lines were assessed using acridine orange (AO) (A-C), an indicator of pH change in apoptotic nuclei, and CED-1::GFP (D-F), a tagged protein expressed in apoptotic cells. (A) Left: Graphic depicting C. elegans stained with AO. Arrows indicate region of meiosis where apoptotic bodies are typically detected. Right: Representative AO-stained gonad from offspring of a live, N2 hermaphrodite exposed to “T&S.” Image is a projection of merged Z-stacks and is cropped to highlight the germ line. Nuclei that accumulate AO (yellow arrowheads) are detected in late pachytene/diplotene of prophase I. (B-C) Apoptotic nuclei of wild-type N2 germ lines were quantified using AO in parents (P0, B) and their offspring (F1, C) in the presence and absence of pesticide treatment. (D) Graphic depicting wild-type C. elegans expressing ced-1::GFP. Green “circles” depict phagocytic nuclei, which are subsequently cleared by apoptosis. (D-E) Quantification of apoptosis using ced-1::GFP-expression in parents (P0, D) and their offspring (F1, E) in the presence and absence of pesticide treatment. Each data point (i.e. shape) corresponds to total number of apoptotic nuclei per gonad arm from a single animal. Number of gonads scored per P0 genotype are as follows: wild-type (N2) control, n = 35; wild-type (N2) “T+S”, n = 42; ced-1::GFP control, n = 45; ced-1::GFP “T+S”, n = 56. Number of gonads scored per F1 genotype are as follows: wild-type (N2) control, n = 64; wild-type (N2) “T+S”, n = 85; ced-1::GFP control, n = 56; ced-1::GFP “T+S”, n = 42. Each corresponding data set is representative of a minimum of 3 independent trials. Significance for all data sets was determined by a two-tailed Mann Whitney test. Horizontal lines correspond to the mean for each data set. Error bars = S.E.M. *denotes p<0.001 and **denotes p≤0.0001. Graphics were created with BioRender.