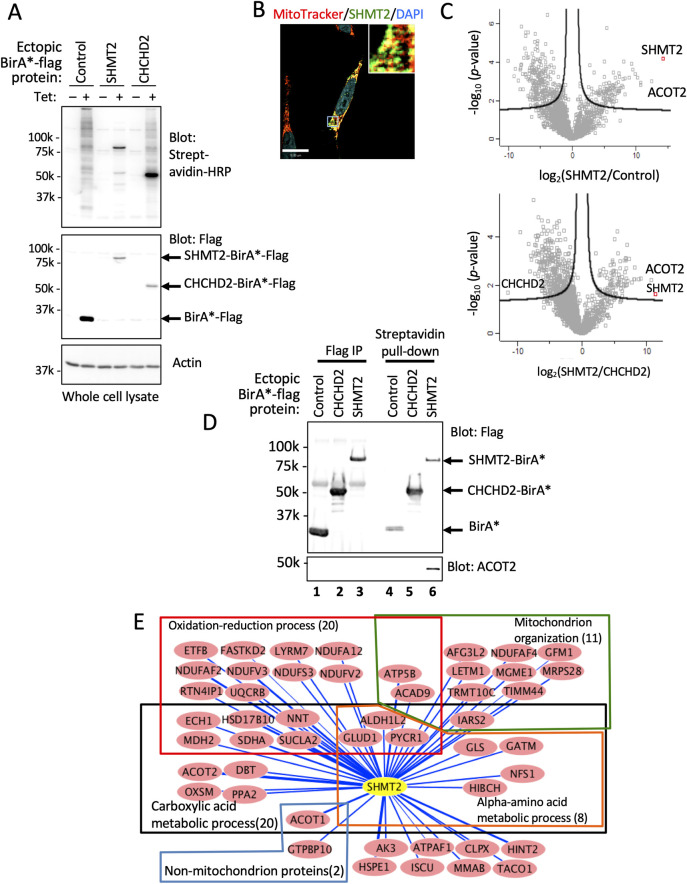
Fig 2. Analysis of SHMT2 interactome by proximity-dependent biotinylation identification (BioID).
(A) Western blot of whole cell lysates from HeLa cells expressing Tet-induced proteins as indicated; proteins have both C-terminal Flag and BirA* tags. (B) Colocalization of MitoTracker Red probe and SHMT2-BirA*-Flag protein. (C) Volcano plot of significance versus fold-change on the y and x axes, respectively, comparing biotinylated proteins identified in SHMT2-BirA-expressing cells and that in two BirA*controls (n = 3). Proteins outside of the lines are significantly different between the indicated two samples. (D) Western blot validation of SHMT2 interacting proteins. Tagged SHMT2 and co-immunoprecipitated partner, ACOT2, was detected with antibodies to SHMT2 and ACOT2. (E) Network analysis of SHMT2 interactome by Cytoscape. Forty-eight proteins significantly different (>32-fold) between ectopic SHMT2 and controls were selected. Proteins and their interactions are shown as nodes and edges. Edges indicate the amount of biotinylated protein in SHMT2-BirA-Flag samples calculated as log10 LFQ signal/protein molecular weight. Gene ontology consortium (http://geneontology.org/) was used for functional classification.