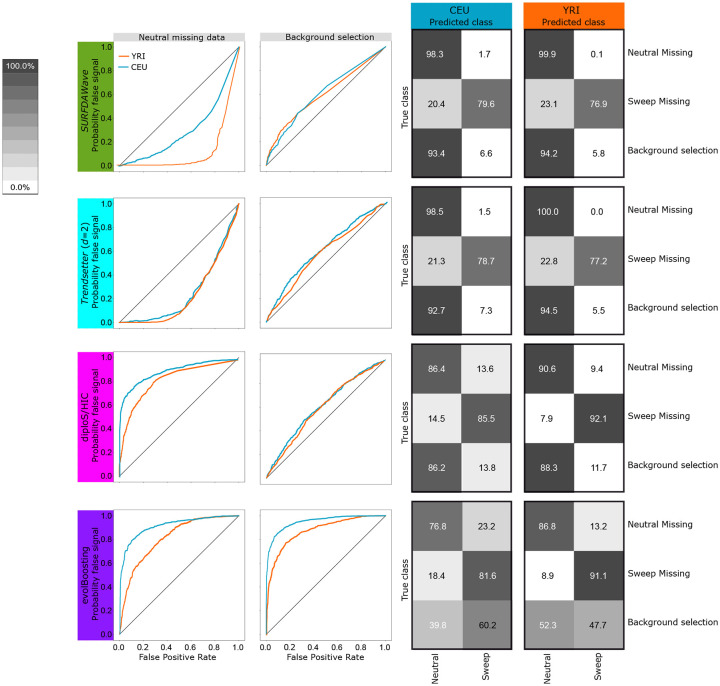
Fig 6. Comparison of method performance under confounding factors (missing data and background selection) when trained with either CEU or YRI demographic histories when SURFDAWave, Trendsetter, diploS/HIC, and evolBoosting are trained to differentiate between sweeps and neutrality.
(Left column) Probability of mis-classifying neutrally-evolving genomic regions missing data as a sweep. Comparing probability of sweep in simulations missing data (probability of false signal) with the probability of any sweep in neutral simulations (false positive rate). (Left middle column) Probability of mis-classifying background selection simulations as sweep. Comparing the probability of a sweep in simulations of background selection (probability of false signal) with probability of sweep in neutral simulations (false positive rate). (Right columns) Confusion matrices showing classification rates when classifying simulations of each class with missing data, and when classifying background selection simulations. Results for both CEU (Right middle column) and YRI (Right column) demographic history. SURFDAWave is trained using Daubechies’ least-Asymmetric wavelets. Optimal γ and and level were chosen through cross validation (see Training the models). Summary statistics , H1, H12, H2/H1, and frequency of the first, second, third, fourth, and fifth most common haplotypes used by both Trendsetter and SURFDAWave.