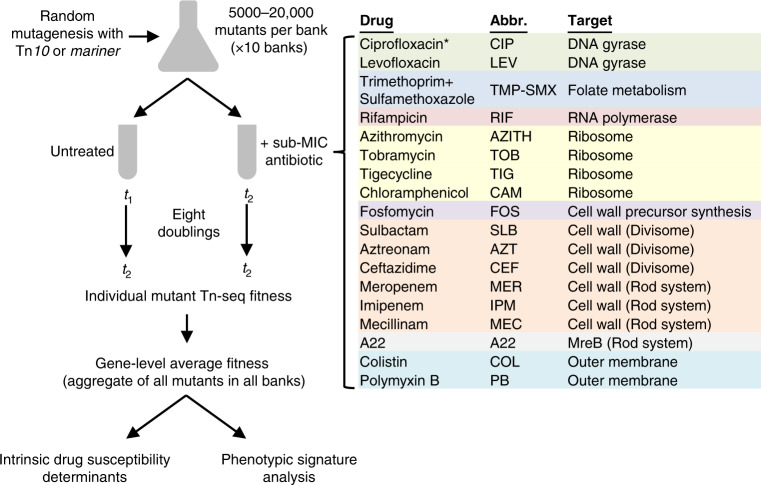
Fig. 1. Genome-wide profiling of gene–antibiotic interactions in A. baumannii by Tn-seq.
Diagram outlines the multiple parallel Tn-seq fitness profiling experiments as described in “Methods”. Sub-MIC drug concentrations used to achieve 20–30% growth rate inhibition are listed in Supplementary Table 1. For each antibiotic, genes contributing to intrinsic defense against each single drug were identified by using significance criteria (Methods). Fitness profiles across all conditions (phenotypic signatures) were then analyzed to identify novel gene relationships and discover multicondition discriminating genes. Drug abbreviations (Abbr.) and targets are listed. *CIP Tn-seq data used in these studies were described previously20.