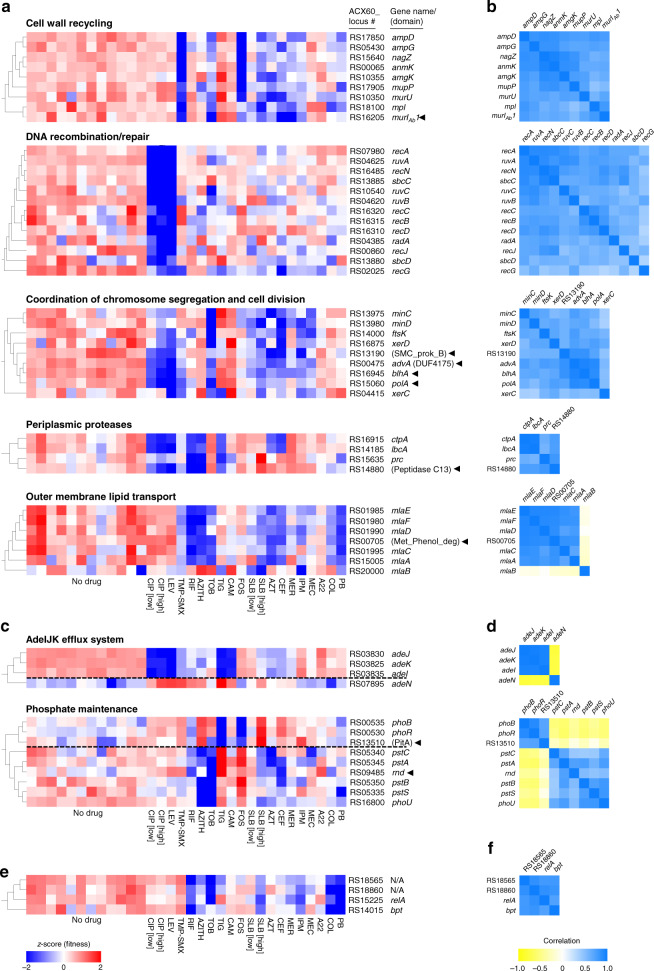
Fig. 2. Genes with interconnected functions show correlated phenotypic signatures.
a–f Genes within a shared functional pathway show relationships in their Tn-seq fitness signatures. a, c Heat maps (using blue-white-red scale at bottom) show normalized Tn-seq fitness in z-scored units for mutants in each gene (rows) grown in distinct conditions (columns). Characterized/annotated genes were first placed into shared functional pathways (Methods). We next identified additional genes (arrowheads), many of which were uncharacterized/unannotated, that correlate with each pathway by using hierarchical clustering with the entire set of A. baumannii phenotypic signatures. Parentheses denote domains identified from NCBI conserved domain database (CDD)46. In c, dashed black lines separate genes with opposing regulatory effects. b, d Heat maps (using yellow-white-blue scale at bottom) show Pearson correlation coefficient (r) matrices measuring relatedness of the corresponding Tn-seq fitness signatures in (a). Positive and negative r indicate correlation and anticorrelation, respectively. e, f Phenotypic signatures correlating with pAB3-encoded ACX60_RS18565. Tn-seq fitness (e) and correlations (f) are shown as above. N/A, no gene name or predicted protein domain. Source data are provided as a Source Data file.