Abstract
Objective
To evaluate the efficacy of sample pooling compared to the individual analysis for the diagnosis of coronavirus disease 2019 (COVID-19) by using different commercial platforms for nucleic acid extraction and amplification.
Methods
A total of 3519 nasopharyngeal samples received at nine Spanish clinical microbiology laboratories were processed individually and in pools (342 pools of ten samples and 11 pools of nine samples) according to the existing methodology in place at each centre.
Results
We found that 253 pools (2519 samples) were negative and 99 pools (990 samples) were positive; with 241 positive samples (6.85%), our pooling strategy would have saved 2167 PCR tests. For 29 pools (made out of 290 samples), we found discordant results when compared to their correspondent individual samples, as follows: in 22 of 29 pools (28 samples), minor discordances were found; for seven pools (7 samples), we found major discordances. Sensitivity, specificity and positive and negative predictive values for pooling were 97.10% (95% confidence interval (CI), 94.11–98.82), 100%, 100% and 99.79% (95% CI, 99.56–99.90) respectively; accuracy was 99.80% (95% CI, 99.59–99.92), and the kappa concordant coefficient was 0.984. The dilution of samples in our pooling strategy resulted in a median loss of 2.87 (95% CI, 2.46–3.28) cycle threshold (Ct) for E gene, 3.36 (95% CI, 2.89–3.85) Ct for the RdRP gene and 2.99 (95% CI, 2.56–3.43) Ct for the N gene.
Conclusions
We found a high efficiency of pooling strategies for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA testing across different RNA extraction and amplification platforms, with excellent performance in terms of sensitivity, specificity and positive and negative predictive values.
Keywords: Pooled analysis, RT-PCR, Sample pooling, SARS-CoV-2, Sensitivity
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has posed an immense challenge for the national health systems of the affected countries. In the absence of a vaccine or effective treatment, molecular diagnosis used to identify transmitting infected patients so they can proceed to isolation to avoid new infections is a crucial tool in containing the pandemic. High demand for testing may be hampered in some scenarios as a consequence of the lack of reagent supplies and their limited production.
The current challenge for SARS-CoV-2 diagnosis is to meet the great demand for testing as we embark on a new era focusing on testing and contact tracing. Clinical laboratories must plan to increase their analytical capacity to face these new public health challenges so that massive analysis can be undertaken to identify all infected persons, proceed with their isolation and trace their contacts. This undoubtedly constitutes a challenge due to the high number of diagnostic processes required and the limited resources available in the face of a disease with a variable incubation period, an uncertain viral dynamic [1,2] and an unknown number of asymptomatic carriers who can unknowingly transmit the infection.
Dorfman in 1943 [3] introduced the strategy into clinical diagnosis of mixing samples together to perform a single test; this strategy has been helpful in correctly identifying all infected individuals using fewer diagnostic tests [4,5]. The diagnosis of SARS-CoV-2 infection is fundamentally based on real-time reverse transcriptase PCR (RT-PCR), which is the reference technique [6,7]. This is a robust technology with high sensitivity and specificity; it has already been used in sample pooling strategies for screening for HIV and hepatitis B and C viruses [[8], [9], [10], [11]], where it has proven to be cost-effective and efficient in surveillance and diagnosis (detection) for a prevalence below 30%, regardless of the population studied. Therefore, the combination of pooled tests and patients at a low risk of infection is considered a practical and effective method to analyse large quantities of samples without compromising precision, especially when it comes to centralized testing models with automated systems.
In SARS-CoV-2 infection, data are limited regarding the best strategy for detecting cases by grouping samples and assessing how they influence the sensitivity of the RT-PCR analysis. It is therefore necessary to investigate the effect of the number of samples, especially with commercially available assays [[12], [13], [14], [15], [16], [17]].
We evaluated the efficacy of sample pooling via a multicentre strategy compared to individual analysis for the detection of coronavirus disease 2019 (COVID-19) by using different commercial platforms available for genomic extraction and amplification by RT-PCR.
Materials and methods
Specimen collection
Between March and May 2020, nasopharyngeal swabs (n = 3519) were collected from patients or health professionals and sent to the virology laboratories of the participating centres (Supplementary Table S1). Nasopharyngeal and pharyngeal swabs were collected at the same time, and both swabs were placed in the same tube with transport media. Several viral transport media were used: Vircell Transport Medium (Vircell, Granada, Spain), δswab Transport Medium (Deltalab, Rubí, Spain) and UTM: Universal Transport (Copan, Brescia, Italy). Samples were processed within the first 24 hours after receipt.
Pooled analysis
Nine or ten individual samples were pooled, and screening was performed using RT-PCR targeting the same target as for individual samples. Pooled testing was performed at each of the participating sites in the study. Pooling was performed by hand, after inactivation of each sample that made up part of the pool. Samples for pooling were selected randomly, according to availability at each site during the study period.
For both individual testing and pooled analysis, samples were inactivated 1:1 in lysis buffer and processed according to the existing methodology in each laboratory (Table 1 ).
Table 1.
Nucleic acid extraction and amplification systems
| Extraction | Amplification | Target | N |
|---|---|---|---|
| Maxwell RSC Viral Total Nucleic Acid (Promega) | Viasure SARS-CoV-2 Real Time PCR (CerTest) | ORF1ab and N gene | 139 |
| m2000sp (Abbot) | Viasure SARS-CoV-2 Real Time PCR (CerTest) | ORF1ab and N gene | 410 |
| MagMAX Viral RNA Isolation Kit (Thermo Fisher Scientific) | TaqMan 2019-nCoV Assay Kit v1 (Thermo Fisher Scientific) | N gene | 70 |
| eMAG (bioMérieux) | TaqMan 2019-nCoV Assay Kit v1 (Thermo Fisher Scientific) | N gene | 190 |
| STARMag (Seegene) | Allplex 2019-nCoV Assay (Seegene) | E, RdRP and N gene | 1910 |
| MagMAX Viral RNA Isolation Kit (Thermo Fisher Scientific) | Light Mix E gene (Roche) | E gene | 240 |
| cobas SARS-CoV-2 test (Roche) | E and ORF1a gene | 560 |
Performance characteristics of pooled analysis
Major discordance was defined as a negative pool result when at least one of the individual samples showed cycle threshold (C t) values of <35 for one or more SARS-CoV-2 genes. Minor discordance occurred when at least one individual sample had C t > 35 in one or two of the SARS-CoV-2 genes assayed and the pool scored negative.
Performance characteristics such as sensitivity, specificity, positive predictive value, negative predictive value and relative efficiency were calculated comparing the individually analysed sample (reference standard) with respect to the sample result analysed within the pool. Statistical analyses were performed by RStudio (https://rstudio.com/) and GraphPad Prism 8 (GraphPad Software, La Jolla, CA, USA) software.
Ethical approval
This study was approved by ‘Comité de Ética de la Investigación con medicamentos de Galicia (CEIm-G)’ review board. Given the deidentified nature of testing, individual patient consent was not required for this study.
Results
The study included 3519 samples from nine different sites in Spain. We analysed all samples individually and also in parallel, pooled into 353 groups of samples (342 pools of ten samples and 11 pools of nine samples). Two hundred forty-one samples (6.85%) were positive.
We found that 253 pools, made up of 2519 samples, were negative (242 pools of ten samples and 11 pools of nine samples); and 99 pools, made up of 990 samples, were positive (99 pools of ten samples). One pool comprising ten samples was invalid. Therefore, our pooling strategy would have saved 2167 (86%) PCR tests. A description of the positive and negative pools is provided in Table 2 .
Table 2.
PCR tests performed with and without pooling strategy
| Result | Pool | Tested individually | Tested with pool strategy |
|---|---|---|---|
| Positive | 99 | 990 | 99 + 990 |
| Negative | 253 | 2519 (2420 + 99) | 253 |
| Invalid | 1 | 10 | 10 |
| Total | 353 | 3519 | 1352 |
Shown are number of samples tested individually and number of samples that would have been tested if pooling strategy would have been run without parallel testing.
Overall, 323 pools with 3219 samples showed concordant results with the individual samples analysed (224 pools with 2229 samples that tested negative, and 99 pools with 990 samples that included at least one positive sample). For 29 pools (290 samples), we found discordant results compared to the individual samples. Minor discordances were found in 22 pools and major discordances in seven. Pools with minor discordances included 24 samples from patients who had a prior positive SARS-CoV-2 test and were submitted for PCR testing at least 20 days afterwards to evaluate RNA clearance (Supplementary Table S2).
When only major discordances were considered, sensitivity, specificity and positive and negative predictive values for pooling were 97.10% (95% confidence interval (CI), 94.11–98.82), 100%, 100% and 99.79% (95% CI, 99.56–99.90) respectively. Accuracy was also 99.80% (95% CI, 99.59–99.92), and the kappa concordant coefficient was 0.984 (Table 3 ).
Table 3.
Performance characteristics of pooled analysis when major discordances were analysed
| Major discordance | Individual test (reference) |
|
|---|---|---|
| No. positive | No. negative | |
| Pool test | ||
| Positive | 234 | 0 |
| Negative |
7 |
3268 |
| Statistic |
Value |
95% CI |
| Sensitivity (%) | 97.10 | 94.11 to 98.82 |
| Specificity (%) | 100.00 | 99.89 to 100.00 |
| PPV (%) | 100.00 | |
| NPV (%) | 99.79 | 99.56 to 99.90 |
| Accuracy (%) | 99.80 | 99.59 to 99.92 |
| Kappa (κ) | 0.984 | |
CI, confidence interval; NPV, negative predictive value; PPV, positive predictive value.
When all discordances were considered for the analysis, sensitivity, specificity and positive and negative predictive values for pooling were 85.48% (95% CI, 80.39–89.67), 100%, 100% and 98.94% (95% CI, 98.57–99.22) respectively. Accuracy was 99.00% (95% CI, 98.62–99.30), and the kappa concordant coefficient was 0.916 (Table 4 ).
Table 4.
Performance characteristics of pooled analysis when all discordances were analysed
| All discordance | Individual test (reference) |
|
|---|---|---|
| No. positive | No. negative | |
| Pool test | ||
| Positive | 206 | 0 |
| Negative |
35 |
3268 |
| Statistic |
Value |
95% CI |
| Sensitivity (%) | 85.48 | 80.39 to 89.67 |
| Specificity (%) | 100.00 | 99.89 to 100.00 |
| PPV (%) | 100.00 | |
| NPV (%) | 98.94 | 98.57 to 99.22 |
| Accuracy | 99.00 | 98.62 to 99.30 |
| Kappa (κ) | 0.916 | |
CI, confidence interval; NPV, negative predictive value; PPV, positive predictive value.
Supplementary Table S3 shows the number and rate of discrepancies across the different tests and targets used at the participating sites. The lowest rate of discrepancies was observed for the Viasure SARS-CoV-2 Real Time PCR (CerTest, Zaragoza, Spain) (n = 3; 0.55%), while the TaqMan 2019-nCoV Assay Kit v1 (Thermo Fisher Scientific, Waltham, MA, USA) showed the highest rate (n = 7; 2.69%).
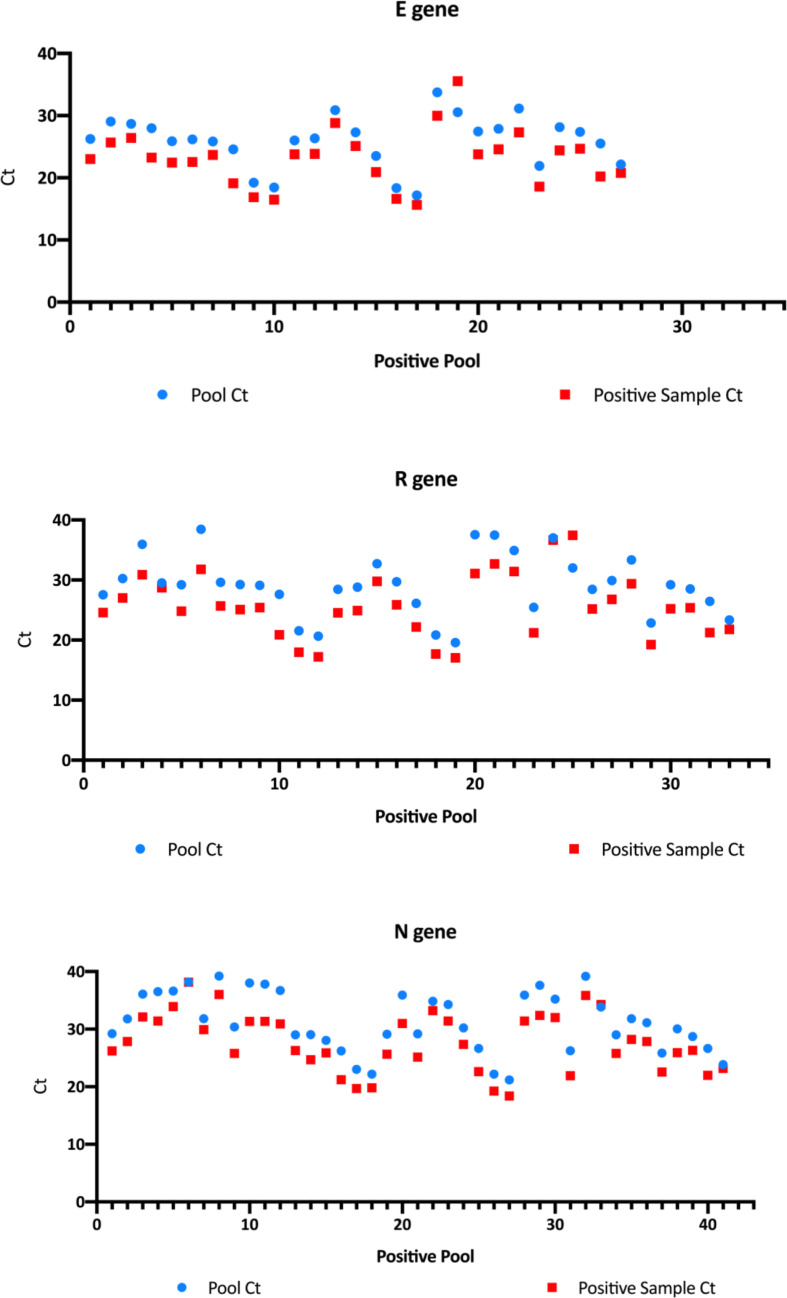
An in-depth analysis of the C t results was performed to check the pooling effect on C t differences. Of the 99 positive pools, 42 were positive pools of ten samples, with one positive sample each. These samples and pools were tested for the E (envelope), RdRP (RNA-dependent RNA polymerase) and N (nucleocapsid) genes (Allplex 2019-nCoV Assay) and analysed by 2019-nCov Seegene (Seoul, South Korea) software. The difference in C t results between pooled and positive sample is provided in Fig. 1 . A median loss was recorded of 2.87 (95% CI, 2.46–3.28) C t for E gene, 3.36 (95% CI, 2.89–3.85) C t for RdRP gene and 2.99 (95% CI, 2.56–3.43) C t for N gene. Interestingly, we found that the most sensitive target after pooling was the N gene (41 pools positive), followed by the RdRP gene (33 pools positive); the E gene (27 pools positive) showed the least sensitivity after pooling samples.
Fig. 1.
Pooling effect on cycle threshold (Ct) results of the specific E, RdRP and N genes from Allplex 2019-nCoV Assay (Seegene, Seoul, South Korea).
Discussion
Pooling strategies had high efficiency for SARS-CoV-2 RNA testing, across different RNA extraction and amplification platforms, with excellent performance in terms of sensitivity, specificity and positive and negative predictive values. We believe that our results may help clinical laboratories respond to the clinical need for SARS-CoV-2 testing.
Although the sample pooling strategy works for other pathogens that are diagnosed by RT-PCR, for SARS-CoV-2 infection, there are still limited data in the literature regarding surveillance and detection strategies by grouping samples [[12], [13], [14], [15], [16], [17]].
We evaluated the efficacy of multicentre sample pooling compared to individual analysis for the detection of COVID-19 by using different commercial platforms available for genomic extraction and amplification by RT-PCR. Within a 6.8% positive rate, we obtained excellent results in terms of sensitivity, specificity and positive and negative predictive values, both in a scenario when only major discordances were considered (97.10%, 100%, 100% and 99.79% respectively) and in a scenario when only minor discordances were counted (85.48%, 100%, 100% and 98.94% respectively).
As expected, and as others have also shown [18], the dilution of samples in our pooling strategy resulted into a median loss of 2.87 C t for E gene, 3.36 C t for RdRP gene and 2.99 C t for N gene. This drop in sensitivity was responsible for most of the discordances found in our study, which were mainly observed for samples with the lowest positivity signals, always with C t values very close to 40 and very frequently in only one gene of the two or three that were included in the tests. Although special attention must be paid to RT-PCR false-negative results [19], it is also known that most of the positive results obtained from just one gene targeted and with C t > 35 correspond to nonviable/noninfectious particles that are still detected by RT-PCR [20]. In addition, false-positive results yielding C t > 35 may also be expected.
Our study's main limitation was the variability in the extraction and amplification methods used, and the number of samples included in the different pools tested; however, this limitation in study design may actually be its main strength, given the consideration that even in this scenario, our results were excellent. When using a sample pooling strategy, it is a priority to determine the group size in which maximum analysis precision is maintained, because as a result of sample dilution, this procedure can decrease the sensitivity of RT-PCR molecular assays [21]. For this reason, before systematically implementing a sample grouping strategy, it is important to consider these characteristics (detection limit, sensitivity and specificity of the test) together with the expected prevalence. In this regard, there are already applications that allow it to be calculated (https://www.chrisbilder.com/shiny/); in addition, an in-depth mathematical analysis of pool testing by Cherif et al. [22] is also available. The main advantages of the pooling strategy are that it allows the use of the same standard protocols of commercial reagents, with no need for additional training, equipment or materials, and consequently it can be implemented immediately to expand the detection and surveillance capabilities of COVID-19. Another limitation to our study is that the samples were pooled by hand. We are currently evaluating the automation of pooling samples, as this is a critical preanalytical step that must be taken before handling the great number of samples that the pooling strategy will allow to process. Result reporting is another critical step that needs to be addressed. Finally, we did not collect information on clinical data or symptoms; we believe that most patients, if not all, would be symptomatic because asymptomatic screening has not been a part of clinical practice in Spain during the pandemic's first wave.
It should be noted that pooling as a screening strategy will not completely eliminate the need for individual diagnostic tests, which will be essential when community transmission intensifies. In our study, even in the setting of a 6.86% prevalence, out of 3519 samples analysed, we would have saved a total of 2167 PCR tests, with great savings in time, costs and personnel. Within the current epidemiologic situation, with significantly decreased prevalence but a high demand for PCR testing, such efficiency measures would help clinical laboratories alleviate their workload in order to prepare for future outbreaks.
In summary, we found that pooling strategies for SARS-CoV-2 RNA testing are highly efficient across different RNA extraction and amplification platforms, with excellent performance in terms of sensitivity, specificity and positive and negative predictive values. Whether pooling is used will depend greatly on SARS-CoV-2 prevalence rather on the fact that patients are asymptomatic (prevalence may be high if community transmission has been reached) or symptomatic. Specific recommendations for pooled testing became available only on 18 July 2020 from the US Food and Drug Administration [23]. Because we believe that our findings may be essential to expand clinical laboratories' capabilities in the near future, we recommend that this strategy be validated in each specific setting of extraction and amplification reagents before its introduction into clinical management of patients, specially to ensure that the sensitivity of the assay—and especially the false-negative rates—are acceptable.
Transparency declaration
All authors report no conflicts of interest relevant to this article.
Editor: F. Allerberger
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2020.09.008.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorfman R. The detection of defective members of large populations. Ann Math Stat. 1943;14:436–440. [Google Scholar]
- 4.Xiong W., Ding J., He Y., Li Q. Improved matrix pooling. Stat Methods Med Res. 2019;28:211–222. doi: 10.1177/0962280217719914. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen N.T., Aprahamian H., Bish E.K., Bish D.R. A methodology for deriving the sensitivity of pooled testing, based on viral load progression and pooling dilution. J Transl Med. 2019;17:252. doi: 10.1186/s12967-019-1992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pascarella G., Strumia A., Piliego C., Bruno F., Del Buono R., Costa F. COVID-19 diagnosis and management: a comprehensive review. J Intern Med. 2020;288:192–206. doi: 10.1111/joim.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tu X.M., Litvak E., Pagano M. Screening tests: can we get more by doing less? Stat Med. 1994;13:1905–1919. doi: 10.1002/sim.4780131904. [DOI] [PubMed] [Google Scholar]
- 9.van Zyl G.U., Preiser W., Potschka S., Lundershausen A.T., Haubrich R., Smith D. Pooling strategies to reduce the cost of HIV-1 RNA load monitoring in a resource-limited setting. Clin Infect Dis. 2011;52:264–270. doi: 10.1093/cid/ciq084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tilghman M.W., Guerena D.D., Licea A., Pérez-Santiago J., Richman D.D., May S. Pooled nucleic acid testing to detect antiretroviral treatment failure in Mexico. J Acquir Immune Defic Syndr. 2011;56:e70–e74. doi: 10.1097/QAI.0b013e3181ff63d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinesha T.R., Boobalan J., Sivamalar S., Subashini D., Solomon S.S., Murugavel K.G. Occult HBV infection in HIV-infected adults and evaluation of pooled NAT for HBV. J Viral Hepat. 2018;25:718–723. doi: 10.1111/jvh.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdalhamid B., Bilder C.R., McCutchen E.L., Hinrichs S.H., Koepsell S.A., Iwen P.C. Assessment of specimen pooling to conserve SARS CoV-2 testing resources. Am J Clin Pathol. 2020;153:715–718. doi: 10.1093/ajcp/aqaa064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deckert A., Bärnighausen T., Kyei N. Pooled-sample analysis strategies for COVID-19 mass testing: a simulation study. Bull World Health Organ. 2020;98:590–598. doi: 10.2471/BLT.20.257188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hogan C.A., Sahoo M.K., Pinsky B.A. Sample pooling as a strategy to detect community transmission of SARS-CoV-2. JAMA. 2020;323:1967. doi: 10.1001/jama.2020.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yelin I., Aharony N., Shaer Tamar E., Argoetti A., Messer E., Berenbaum D. Evaluation of COVID-19 RT-qPCR test in multisample pools. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres I., Albert E., Navarro D. Pooling of nasopharyngeal swab specimens for SARS-CoV-2 detection by RT-PCR. J Med Virol. 2020 doi: 10.1002/jmv.25971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wacharapluesadee S., Kaewpom T., Ampoot W., Ghai S., Khamhang W., Worachotsueptrakun K. Evaluating the efficiency of specimen pooling for PCR-based detection of COVID-19. J Med Virol. 2020 doi: 10.1002/jmv.26005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohse S., Pfuhl T., Berkó-Göttel B., Rissland J., Geißler T., Gärtner B. Pooling of samples for testing for SARS-CoV-2 in asymptomatic people. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woloshin S., Patel N., Kesselheim A.S. False negative tests for SARS-CoV-2 infection—challenges and implications. N Engl J Med. 2020;383:e38. doi: 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- 20.Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aragón-Caqueo D., Fernández-Salinas J., Laroze D. Optimization of group size in pool testing strategy for SARS-CoV-2: a simple mathematical model. J Med Virol. 2020 doi: 10.1002/jmv.25929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cherif A., Grobe N., Wang X., Kotanko P. Simulation of pool testing to identify patients with coronavirus disease 2019 under conditions of limited test availability. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.US Food and Drug Administration (FDA) FDA news release; 2020. Coronavirus (COVID-19) update: FDA issues first emergency authorization for sample pooling in diagnostic testing.https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-first-emergency-authorization-sample-pooling-diagnostic 18 July Available at: [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.