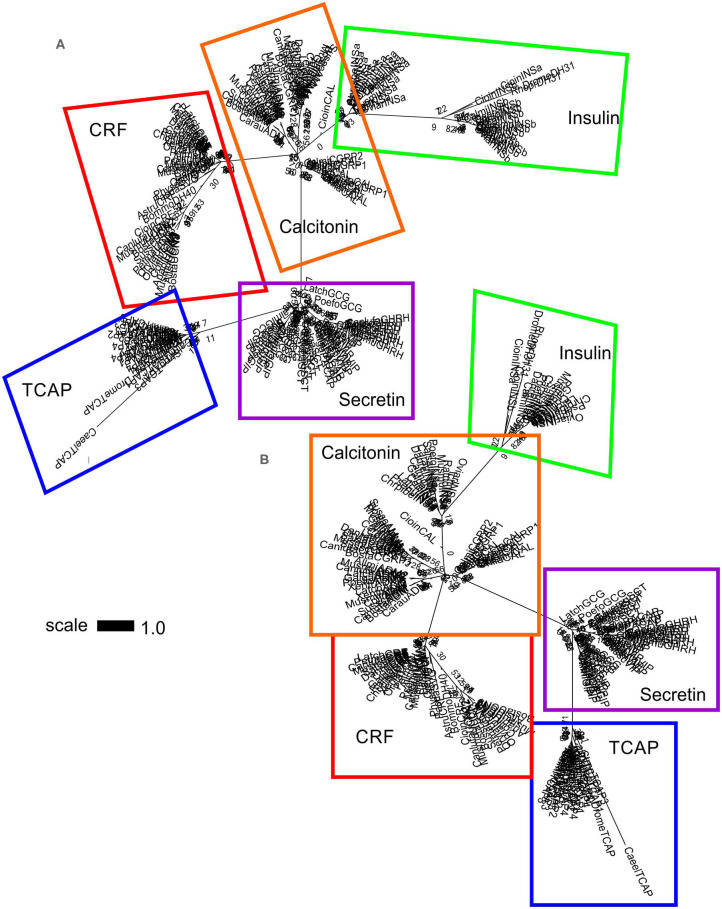
Figure 6.
Phylogenetic analysis of insulin, calcitonin, CRF, secretin, and TCAP mature peptides. The trees are represented as (A) unrooted and (B) rooted to TCAP. Each family is highlighted with a different color: CRF (red), calcitonin (orange), insulin (green), secretin (purple), and TCAP (blue). Analysis was conducted using the maximum likelihood method based on the Whelan and Goldman model (lnL = −1781.0007; +G, parameter = 23.5912) (41). Initial trees for the heuristic search were obtained by applying the NJ method to a matrix of pairwise distances estimated using a JTT model. Branch lengths represent the number of substitutions per site, with the tree shown to scale. Bootstrap analysis involved 1,000 replicates. Calcitonin family: CALC, calcitonin; CGRP1, calcitonin-gene-related peptide 1; CGRP2, calcitonin-gene-related peptide 2; AM, amylin; ADM, adrenomedullin; ADM2, adrenomedullin 2; INSa, insulin A chain; INSb, insulin B chain; CRF family: CRF, corticotropin-releasing factor; TCN, teleocortin; UCN, urocortin; UCN2, urocortin 2; UCN3, urocortin 3; UI, urotensin; SVG, sauvagine; DH, diuretic hormone; Secretin family: SCT, secretin; GHRH, growth hormone releasing hormone; GIP, gastric inhibitory peptide; GCG, glucagon; PACAP, pituitary adenylate cyclase-activating peptide; VIP, vasoactive intestinal peptide; Outgroup: TCAP, teneurin C-terminal associated peptide. The scale bar indicates the level of magnification for the tree.