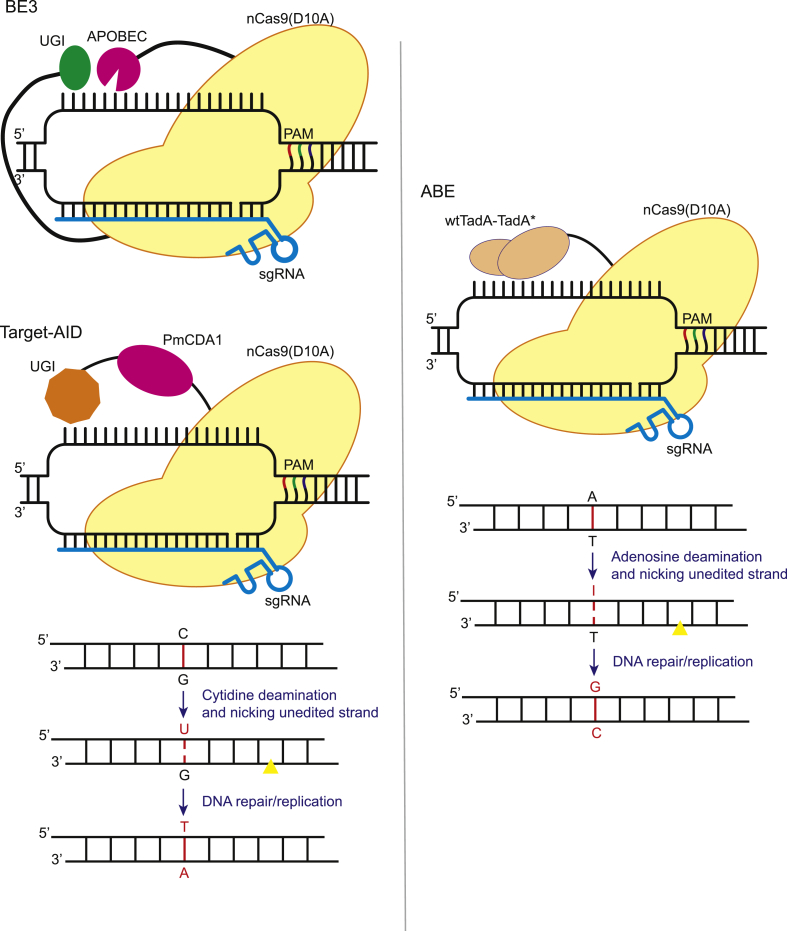
Fig. 1.
Deaminase based Cas9 base editing. A schematic model of deaminase based Cas9 base editing systems: cytosine base editors (CBE, for example, BE3 & Target-AID) and adenine base editor (ABE). CBE consists of rat apolipoprotein B mRNA editing enzyme (APOBEC), and uracil glycosylase inhibitor (UGI) fused to N and C terminus of nCas9(D10A), respectively. Target-AID consists of activation-induced cytosine deaminase ortholog PmCDA1 and UGI fused to N terminus of nCas9(D10A). CBE involves deamination of cytosine by the deaminase which converts it into uracil, making U•G wobble base. UGI prevents its conversion back to C. Mismatch repair machinery (MMR) recognizes it forming U•A which is then converted to T•A by replication machinery leading C•G-to-T•A substitution. ABE consists of heterodimeric wtTadA-TadA* fused to nCas9(D10A). ABE performs deamination converting T•A-to-T•I which is then recognized by DNA repair and replication machinery and converted to C•G base pair (Yellow triangle: nick site).