Abstract
Global warming affects primary producers in the Arctic, with potential consequences for the bacterial community composition through the consumption of microalgae-derived dissolved organic matter (DOM). To determine the degree of specificity in the use of an exudate by bacterial taxa, we used simple microalgae–bacteria model systems. We isolated 92 bacterial strains from the sea ice bottom and the water column in spring–summer in the Baffin Bay (Arctic Ocean). The isolates were grouped into 42 species belonging to Proteobacteria, Bacteroidetes, Actinobacteria and Firmicutes. Forty strains were tested for their capacity to grow on the exudate from two Arctic diatoms. Most of the strains tested (78%) were able to grow on the exudate from the pelagic diatom Chaetoceros neogracilis, and 33% were able to use the exudate from the sea ice diatom Fragilariopsis cylindrus. 17.5% of the strains were not able to grow with any exudate, while 27.5% of the strains were able to use both types of exudates. All strains belonging to Flavobacteriia (n = 10) were able to use the DOM provided by C. neogracilis, and this exudate sustained a growth capacity of up to 100 times higher than diluted Marine Broth medium, of two Pseudomonas sp. strains and one Sulfitobacter strain. The variable bioavailability of exudates to bacterial strains highlights the potential role of microalgae in shaping the bacterial community composition.
This article is part of the theme issue ‘The changing Arctic Ocean: consequences for biological communities, biogeochemical processes and ecosystem functioning'.
Keywords: Arctic Ocean, diatoms, dissolved organic exudates, biodegradation, bacterial isolation, bacterial diversity
1. Background
Phytoplankton are major contributors to the dissolved organic matter (DOM) pool in marine environments by different processes such as grazing by zooplankton, viral lysis, autolysis and exudation [1]. Dissolved organic exudates from phytoplankton are considered as labile and semi-labile molecules composed of sugars (monosaccharides, oligosaccharides and polysaccharides), nitrogen compounds (amino acids, polypeptides or proteins), lipids (fatty acids) and organic acids (vitamins) [2]. On average, DOM exudation accounts for 10–20% of total primary production in the marine environment and for 2–10% (average 5%) for exponentially growing cells [1]. However, different environmental factors may increase the percentage extracellular release (PER), such as suboptimal light intensity and temperature, nutrient limitation and high pCO2 [2–4].
Phytoplankton primary production and PER are both affected by the unprecedented changes occurring in the Arctic Ocean over the past several decades due to climate variations and human activities. The Arctic air temperatures have increased by 2.7°C on the annual scale over the past 47 years (1971–2017), which is twice as fast as in the rest of the Northern Hemisphere [5]. This global warming led to dramatic modifications in the Arctic sea ice, both in the rates and the magnitude of change in extent, area, thickness and spatial distribution [5]. This results in a greater transmission of light through the ice and into newly formed open ocean areas [6]. It is expected that the increase in light intensity in the water column will promote phytoplankton primary production and biological productivity throughout the Arctic Ocean [6,7]. In addition, while phytoplankton blooms were until recently considered negligible in the water column under the ice pack [8,9], recent studies have shown the existence of blooms under the sea ice in the marginal ice zone [10–13]. Diatoms are an important component of the primary production in both pelagic and under sea ice environments during spring blooms in the Arctic Ocean [14–17]. However, measurements of PER in the Arctic Ocean are scarce. Engel et al. [3] measured PER values between 11 and 23% during mesocosm experiments conducted from June to July in Kongsfjorden, Svalbard.
Bacteria are key organisms in carbon cycling in aquatic ecosystems, acting as a sink (mineralization of dissolved organic carbon (DOC) to CO2) or as a link (production of biomass which can be transferred through the microbial food web) [18]. Roughly 50% of carbon fixed by primary production in the ocean passes through bacteria [19]. The quality of the DOM produced by phytoplankton constitutes an important regulator of both the metabolism and composition of bacterial communities [20]. Interactions between heterotrophic bacteria and diatoms are complex [21]. On the one hand, bacteria exert positive (e.g. production of vitamins) or negative effects (e.g. production of algicidal metabolites) on diatom growth [22,23] and can also modify the composition of the diatom exudates [24]. These effects appear to be diatom species-specific, resulting in compositional shifts of diatom communities when different diatom species were grown together [25]. On other hand, DOM released during a diatom bloom can reshape the bacterial community by favouring some bacterial groups like Roseobacter, Sulfitobacter and Flavobacteriaceae that are more adapted to the rapid use of substrates released by diatoms [20]. Consequently, the modification of the primary producers in the Arctic Ocean induced by global warming has potential consequences for the bacterial community composition through the consumption of microalgae-derived DOM [26]. Some experimental studies have shown that bacterial community composition changes are dependent on microalgae species [27], whereas other studies observed a weak bacterial selectivity of exudates produced by specific algae species [28,29]. However, extrapolation of these results is complicated by temporal [30] and spatial [31] changes in the bacterial response to the same DOM exudate. Additionally, these different studies have been performed on natural bacterial communities where complex interactions can occur (e.g. competition, co-metabolism and antibiosis). By contrast, the capacity of distinct bacterial strains to use the exudates from phytoplankton is not well known. Only a simplified approach, combining one bacterial strain with the excretion product of one phytoplankton species, allows us to decipher more precisely the intrinsic capacity of bacteria to use diatom DOM. This type of study has thus far not been carried out, and it complements those with natural microbial consortia.
To determine the degree of specificity in the use of an exudate by bacterial taxa, we used simple microalgae–bacteria model systems. We first identified different bacterial strains isolated from the sea ice bottom and water column during the spring phytoplankton bloom in the Baffin Bay. The fieldwork for the isolation of the bacterial strains was carried out as part of the Green Edge project (http://greenedge-expeditions.com/) with its main objective being to study the phytoplankton development in spring [32]. The study site, which is located in a marginal sea (360 m depth) of the Baffin Bay, is covered by a seasonally changing layer of ice. The ecological relevance of the bacterial strains isolated was evaluated by comparing them with the environmental 16S rRNA sequences retrieved during the same sampling period [33]. Then, we followed the capacity of the bacterial strains to grow on the exudates produced by two Arctic diatoms, Chaetoceros neogracilis and Fragilariopsis cylindrus, characteristics of planktonic and sea ice environments, respectively [17,34,35]. This experimental design allowed us to study the link between the phylogeny of isolated bacteria as well as their origin of their isolation and their capacity to degrade each type of exudate.
2. Methods
(a). Sampling
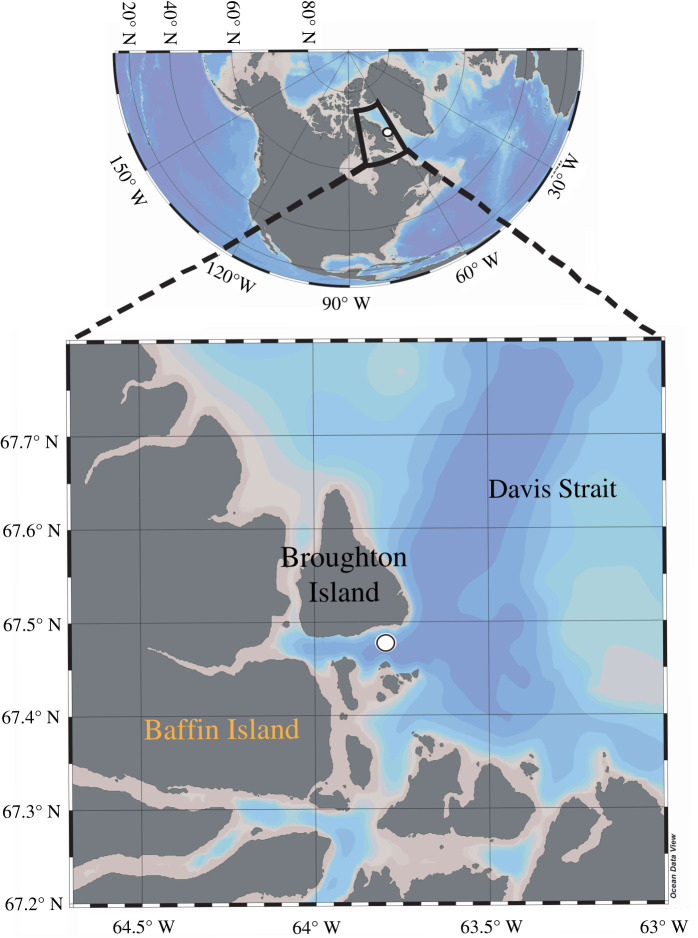
A sampling campaign was conducted from 20 April to 27 July 2016, on an ice camp located near Qikiqtarjuaq (Nunavut), in the Canadian Arctic at 67°28.784′ N, 063°47.372′ W (figure 1). This period was chosen in order to capture the dynamics of the sea ice algae and phytoplankton spring bloom from initiation to termination. Water sampling was carried out through a 1 × 1 m hole in the ice pack shielded by a tent. Water samples were collected every week at 2 and 40 m (or 60 m) depths using a Niskin bottle. The bottom of the first-year sea ice (first 10 cm) was subsampled from sea ice cores collected using a manual ice corer and melted at 4°C.
Figure 1.
Location of Qikiqtarjuaq Ice Camp (67°28.784′ N, 063°47.372′ W). The position is shown with a white dot (courtesy of R. Amiraux). (Online version in colour.)
(b). Bacterial strain isolation and identification
An aliquot of each sample (0.5 ml) was placed in cryotubes containing 500 µl of 70% sterile glycerol and stored at −80°C before isolation at the laboratory. After thawing, 100 µl of each sample was spread in triplicate on Marine Agar medium ([36], electronic supplementary material, table S1), and the Petri dishes were incubated in the dark at 4°C to allow the development of aerobic heterotrophic psychrotolerant marine bacteria. At regular intervals (one, two and three weeks), the colonies were counted and categorized based on their morphological characteristics (morphotype). Representatives of each morphotype were selected for isolation by repeated streaking. Finally, we isolated 92 bacterial strains in total.
After isolation and purification, each strain was cultivated in Marine Broth (MB) medium ([36], electronic supplementary material, table S1) at 4°C under agitation and darkness to identify the strains by partial sequencing of the 16S rRNA. Bacterial cells corresponding to each strain were recovered by centrifugation (3 min at 14 000 g) from 2 ml of MB culture. The DNA was then extracted using the Wizard® Genomic DNA Purification Kit (Promega) following the manufacturer's recommendations. The DNA was amplified by the PCR using the primer pair 27f/1492r to target the 16S rRNA gene [37,38]. The complete PCR mixture used contained ultrapure water (3.2 µl), KAPA2G mix (5 µl), forward primer (0.4 µM final concentration), reverse primer (0.4 µM final concentration) and DNA template (1 µl) for a total reaction volume of 10 µl. The PCR amplification conditions used were as follows: 5 min at 95°C; 35 cycles of 15 s at 95°C, 15 s at 50°C and 15 s at 72°C; 5 min at 72°C and finally holding at 4°C in a Veriti™ Thermal Cycler (Applied Biosystems). The PCR products, previously controlled by electrophoresis, were then purified using an AmpliClean™ Magnetic Bead-Based PCR Cleanup Kit (Nimagen) according to the manufacturer's instructions. DNA was then sequenced by the Sanger method [39]. To do this, a sequence reaction was performed on the samples using the 907r primer [40]. The complete PCR mixture used contained ultrapure water (5.75 µl), BDT buffer (1.75 µl), BigDye Terminator v.3.1 kit (0.5 µl), primer (3.2 µM, 1 µl) and DNA matrix (1 µl) for a total reaction volume of 10 µl. The PCR amplification conditions used were as follows: 40 cycles of 10 s at 95°C, 5 s at 50°C and 2.5 min at 60°C in a Veriti™ Thermal Cycler (Applied Biosystems). The PCR products were cleaned with the D-pure DyeTerminator Cleanup Kit (Nimagen) before being sequenced using a Sanger 16 capillary sequencer AB3130XL (Applied Biosystems).
(c). Sequence analysis
Partial 16S rRNA gene sequences were trimmed, quality controlled and dereplicated using the package Staden-GAP4 [41]. For bacterial strain identification, each FASTA file was uploaded in Ez Taxon-e [42] and compared with the cultured bacterial strain database using the Basic Local Alignment Search Tool (BLAST). The sequences were aligned using the MUSCLE multiple alignment method [43]. A hierarchical likelihood-ratio test was performed to determine the most appropriate substitution model for our dataset. We used default parameters for these different tools. Finally, a phylogenetic tree was constructed from the model used according to the maximum-likelihood (ML) criterion. To study the robustness of the tree nodes, a bootstrap analysis was performed with 1000 repetitions. All sequence processing was performed using Mega X v.10.0.1 software [44]. Sequences were deposited in GenBank (NCBI) under the following numbers: MK 224724–224815 for the 92 bacterial strains isolated during this study and MK 217878–217881 for the four bacterial strains isolated in August 2009 at 3 m depth in the Beaufort Sea and used in the microplate growth assay (see below). The strains were registered in the Banyuls Bacterial Culture Collection (BBCC) (https://collection.obs-banyuls.fr/index.php).
(d). Culture of Arctic microalgae and recovery of exuded dissolved organic matter
Two diatoms from the Roscoff Culture Collection (RCC) of the Roscoff Biological Station (http://roscoff-culture-collection.org/) were used: the pelagic strain RCC 2278 Chaetoceros neogracilis (Mediophyceae, [45]) and the ice strain RCC 4289 Fragilariopsis cylindrus (Bacillariophyceae, Grunow ex Cleve). Both strains were isolated from the Arctic Ocean. Cultures were not axenic. These diatoms were cultured at 4°C with a continuous white light intensity of 10 µE in K/2 medium + Si [46] for C. neogracilis and L1 medium [47] for F. cylindrus. The composition of these different media is given on the website of the RCC (http://roscoff-culture-collection.org/). The diatoms were then transferred to a modified K culture medium in which (i) tris(hydroxymethyl)aminomethane (Tris-Base), Na2-Glycerophosphate (C3H7O6PNa2, 5–6 H2O) and ethylenediaminetetraacetic acid (EDTA) as organic molecules were removed, (ii) ammonium was removed to reduce bacterial growth during culture and (iii) iron concentration was reduced to 11.7 µM to limit iron precipitation due to the elimination of EDTA. The culture flasks containing 200 ml of modified K medium were incubated under the same conditions as before. Cell growth was measured by flow cytometry (BD Accuri C6 Plus, Becton Dickinson). The samples were fixed with glutaraldehyde (0.25% final) and stored at −20°C before analysis. The diatoms were detected by chlorophyll fluorescence and side-angle light-scattering properties using blue laser (488 nm). Maximum growth rates (μmax) in the exponential phase were calculated using cell counts according to the equation: μmax = ln (N2/N1)/(t2 – t1), where μ represents doublings (d−1); N represents algal abundance ml−1 and t represents time (d−1). Bacteria were also counted after staining with SYBR Green (Invitrogen, 0.05% final) for 10 min in the dark before being run by flow cytometry. The bacteria were detected by green fluorescence and side-angle light-scattering properties using blue laser (488 nm). Flow cytometric results were processed using the BD CSampler™ C6 v.1.0.241.21 software (Becton Dickinson). Results were analysed with the FlowJo v.7.6.1 (FlowJo LLC) software.
The diatom DOM was harvested after 40–44 days of cultivation, corresponding to a prolonged stationary phase. This time period was chosen to ensure the collection of a sufficient amount of organic exudates for the subsequent bioassay experiments. Therefore, triplicate culture flasks for each strain were pooled and then centrifuged for 15 min at 5166 g at 4°C. The supernatant was then filtered through two GF/F filters (0.7 µm, 47 mm, Whatman) using a glass vacuum flask. To avoid any organic contamination, the filters and glassware were previously combusted at 450°C for 6 h. The filtrate of each strain was stored at −20°C in Nalgene vials, previously washed with 10% hydrochloric acid (HCl) and then rinsed with Milli-Q® water. Inorganic nutrients and DOC were analysed at the beginning and at the end of the culture for each diatom strain (see below). Particulate organic carbon (POC) and dissolved and total saccharides were analysed at the end of the culture (see below).
(e). Chemical analyses
Samples for POC (10 ml aliquots of culture in duplicate) were filtered onto pre-combusted (450°C for 6 h) GF/F filters, which were then stored at −20°C. Before analysis, filters were placed in a desiccator for 48 h at room temperature. Samples were analysed with a Carlo Erba Instruments EA1108 elemental analyser using an acetanilide standard as a reference. Samples (10 ml aliquots of culture in duplicate) for nitrate (NO3−), phosphate (PO43−) and silicate (Si(OH)4) were stored at −20°C before analysis on a nutrient autoanalyzer (SEAL Analytical AA3HR). DOC samples (20 ml aliquots of culture in duplicate) were filtered through two overlaid combusted (450°C, 6 h) 25 mm GF/F filters. The filtrate was transferred into combusted glass tubes, poisoned with 85% H3PO4, closed with Teflon-lined screw caps and stored in the dark at room temperature until analysis. Samples (65 ml) for dissolved and total saccharides analyses were stored at −20°C.
Carbohydrates were determined in the filtrates of each strain by the TPTZ method [48,49]. This method detects the following categories of monosaccharides: hexoses (fructose, galactose, glucose and mannose), pentoses (ribose, arabinose and xylose), deoxy sugars (fucose and rhamnose) and uronic acids (glucuronic, galacturonic and mannuronic acids). Dissolved-free monosaccharides (DFCHO) were determined without acid hydrolysis. Total dissolved monosaccharides (TDCHO) were determined after acid hydrolysis using 1 M HCl (final concentration) at 100°C for 20 h [50]. Hydrolysis was stopped after placing the tubes in an ice bath for 5 min. Combined monosaccharides (i.e. dissolved polysaccharides or DPCHO) were then calculated as the difference between TDCHO and DFCHO. A standard calibration curve was obtained by the analysis of five glucose standards spanning a concentration of 0.2–2 mg l−1. The response was linear over the above concentration range (R2 = 0.999), and monosaccharides concentrations were converted to C-equivalents by using a molar conversion factor of 6.
(f). Microplate assay to measure bacterial growth with microalgae exudates
We studied the ability of 40 different bacterial strains, each corresponding to a different species, to degrade the DOM exuded by the two Arctic diatom strains. Among these bacterial strains, 36 were selected from the 92 bacterial strains isolated within the Green Edge project and four additional strains, isolated in the Beaufort Sea at 3 m depth in August 2009 [51], were also selected. Bacterial strains were cultured in an MB at 4°C in the dark for 3–5 days, then 2 ml of culture were centrifuged at 8000 g for 5 min at 4°C. The cell pellets were washed three times with sterile artificial seawater [52], with a centrifugation cycle between each wash as before, in order to remove the DOM contained in the culture medium. The inoculum of each strain was resuspended in artificial seawater, and the cell concentration was determined by flow cytometry as described above (Cytoflex, Beckman & Coulter).
Bacterial growth assays were conducted in 24-well microplates. Each well contained 2 ml of bacterial suspension adjusted to 2 × 105 cell ml−1 in artificial seawater. Dissolved organic exudates were added in triplicate at a final concentration of 375 µM DOC. NO3− and PO43− were adjusted at 184 and 16 µM final concentrations, respectively, to avoid any effect of inorganic nutrient concentration differences between the two exudates tested. Positive controls were prepared in triplicate for each strain by adding diluted MB (1/560) at the same DOC concentration than for exudates. Negative controls were also prepared in triplicate for each strain by adding a volume of artificial seawater equivalent to those of exudates and diluted MB. Microplates were incubated at 4°C in the dark with a slight agitation. Bacterial growth was monitored every 2 days for 6 days by flow cytometry using the same protocol described above, taking 50 or 100 µl samples in each well. Preliminary assays were performed with different MB dilutions to confirm that bacterial growth was measurable at this time scale of incubation with a concentration of 375 µM DOC (electronic supplementary material, figure S1).
To estimate the growth capacity of the bacterial strains, we used a first index that is the ratio of the maximum cell abundance to the initial cell abundance: , where [B]T0 corresponds the initial cell concentration and [B]max to the maximum cell concentration. We considered that growth was positive when this index was greater than or equal to 4, which corresponds to two cell divisions.
To specify the growth capacity of the strains to use exudates comparatively to the MB, a second index was calculated that considers the intrinsic growth of each strain. For this purpose, we reported the growth obtained with exudates compared with that obtained with diluted MB (positive control). The mathematical formula for this index was:
With [B]T0 exudate the initial cell concentration in the exudate, [B]max exudate the maximum cell concentration with the exudate, [B]T0 MB the initial cell concentration with the diluted MB and [B]max MB the maximum cell concentration obtained with the diluted MB. This index was calculated only for strains showing positive growth with index 1.
3. Results
(a). Viable counts and cultivable bacterial diversity
Percentages of cultivability varied between 0.002 and 4.3% (table 1). The percentages of cultivability were not significantly different between the different habitats even if the sea ice samples were characterized by much higher concentrations of chlorophyll a compared with the samples from the water column (table 1). No temporal trend was observed in the percentages of cultivability.
Table 1.
Cultivable bacteria counts for sea ice bottom and water column samples collected at an ice camp (67°28.784′ N, 063°47.372′ W) located near Qikiqtarjuaq (Nunavut) in the Baffin Bay. CFU: colony-forming units; ND: no data.
| habitat | sampling date (d/m/y) | Julian day | temperature (°C) | salinity (psu) | Chla (μg l−1) | CFU (102 ml−1) | total bacteria (105 ml−1) | CFU/ total (%) | number of isolated strains |
|---|---|---|---|---|---|---|---|---|---|
| sea ice | 09/05/2016 | 130 | ND | ND | 30 | 17.2 | 1.08 | 1.59 | 8 |
| sea ice | 23/05/2016 | 144 | ND | ND | 95.87 | 8.7 | 4.87 | 0.18 | 7 |
| sea ice | 30/05/2016 | 151 | ND | ND | 164.88 | 0.1 | 5.26 | 0.00 | ND |
| sea ice | 06/06/2016 | 158 | ND | ND | 61.89 | 0.9 | 4.6 | 0.02 | ND |
| sea ice | 20/06/2016 | 172 | ND | ND | 138.01 | 17.5 | ND | ND | 8 |
| sea ice | 27/06/2016 | 179 | ND | ND | 39.68 | 27.1 | 6.24 | 4.35 | 5 |
| 2 m | 09/05/2016 | 130 | −1.73 | 32.22 | 0.06 | 11.5 | 2.01 | 0.57 | 7 |
| 2 m | 16/05/2016 | 137 | −1.74 | 32.24 | ND | 20.8 | 2.62 | 0.80 | ND |
| 2 m | 23/05/2016 | 144 | −1.73 | 32.22 | 0.07 | 32.6 | 2.64 | 1.23 | 6 |
| 2 m | 30/05/2016 | 151 | −1.71 | 32.26 | 0.16 | 15.5 | 2.47 | 0.63 | ND |
| 2 m | 06/06/2016 | 158 | −1.7 | 32.28 | 0.88 | 12.7 | 2.38 | 0.53 | ND |
| 2 m | 13/06/2016 | 165 | −1.69 | 32.29 | 0.36 | 23.3 | 2.71 | 0.86 | 6 |
| 2 m | 20/06/2016 | 172 | −1.62 | 32.16 | 0.4 | 19.5 | 3.51 | 0.56 | 6 |
| 2 m | 27/06/2016 | 179 | −1.51 | 32.1 | 1.06 | 14.5 | 5.09 | 0.29 | 7 |
| 2 m | 11/07/2016 | 193 | −1.29 | 31.6 | 1.44 | 4.6 | 6.81 | 0.07 | 6 |
| 40 m | 09/05/2016 | 130 | −1.67 | 32.28 | 0.13 | 5.7 | 2.09 | 0.27 | 8 |
| 40 m | 16/05/2016 | 137 | −1.68 | 32.35 | ND | 2.9 | 2.35 | 0.12 | ND |
| 40 m | 23/05/2016 | 144 | −1.69 | 32.27 | 0.06 | 3 | 2.43 | 0.12 | 4 |
| 40 m | 30/05/2016 | 151 | −1.65 | 32.36 | 0.05 | 0.7 | 2.38 | 0.03 | ND |
| 40 m | 06/06/2016 | 158 | −1.68 | 32.31 | 0.08 | 3.8 | 2.56 | 0.15 | ND |
| 40 m | 13/06/2016 | 165 | −1.63 | 32.44 | 0.06 | 13.2 | 2.59 | 0.51 | 5 |
| 60 m | 27/06/2016 | 179 | −1.58 | 32.51 | 0.32 | 10.5 | 3.16 | 0.33 | 6 |
| 60 m | 11/07/2016 | 193 | −1.57 | 32.38 | 0.59 | 1.6 | 3.62 | 0.04 | 3 |
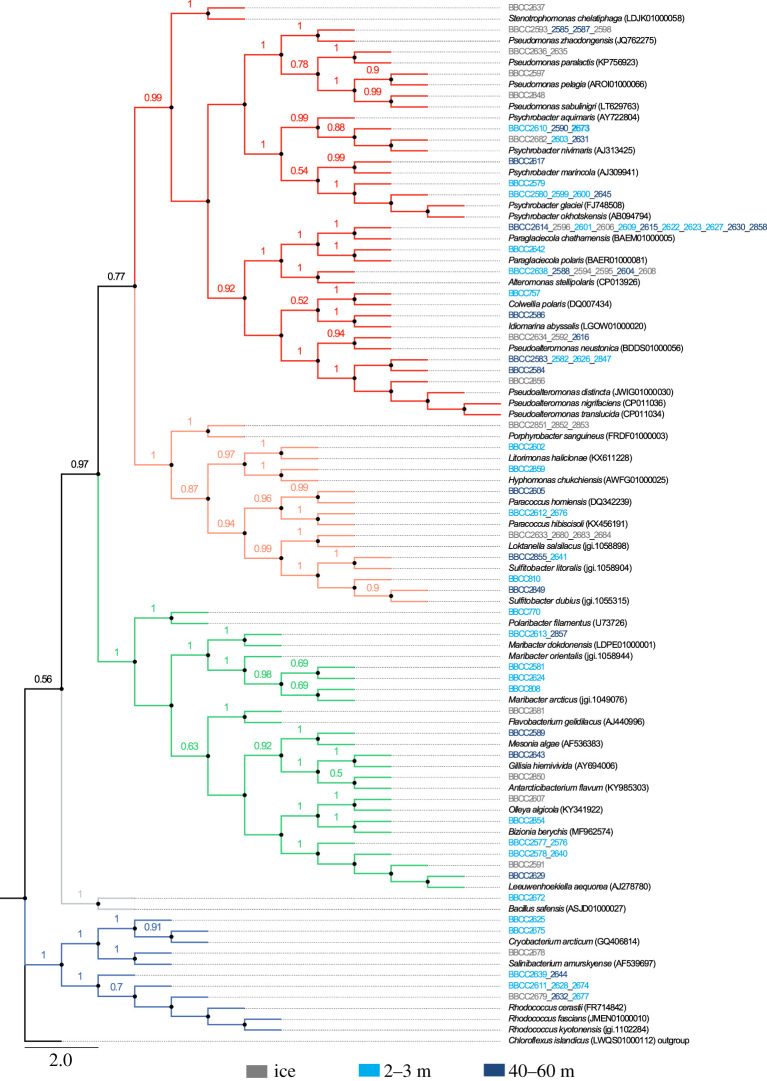
Sequence analysis of the 92 isolated bacterial strains allowed the identification of 42 putatively different species, according to similarity criterion ≥99% (1% divergence) for the 16SrRNA gene sequence, belonging to four major phyla (figure 2; electronic supplementary material, figure S2A). The percentages of similarity of the 16S rRNA gene between the isolated strains and the closest reference sequences were between 95.72 and 100%. The bacterial strains were mostly similar to species previously isolated from polar environments. They belong primarily to Proteobacteria (70% of the 92 sequences identified) and more particularly to Gammaproteobacteria, which represented 53% of all the sequences (electronic supplementary material, figure S2). The remaining Proteobacteria were Alphaproteobacteria (16% of the total sequences). The two other phyla were Bacteroidetes and Actinobacteria, representing 16 and 13% of the sequences, respectively. Finally, one bacterial strain belonging to the Firmicutes phylum was isolated.
Figure 2.
Phylogenetic tree of bacterial strains isolated from ice camp samples and closely related species obtained from the GenBank. The codes indicate the environmental Arctic strains isolated (table 3). The GenBank accession number follows specific names of the closely related species found in the database. The phylogenetic tree was made using the maximum-likelihood method (ML). The phylogeny obtained was statistically analysed by the bootstrap method, with 1000 repetitions (values greater than 0.5 are shown at each node). In red: Gammaproteobacteria; in orange: Alphaproteobacteria; in green: Flavobacteriia; in blue: Actinobacteria; in grey: Firmicutes. The Chloroflexus islandicus sequence has been used as outgroup to root the tree. Scale bar represents 2% estimated substitutions. (Online version in colour.)
When the phylogeny of the bacterial strains was analysed according to the sampling habitat, we observed only slight modifications in the relative proportion of each group (electronic supplementary material, figure S2B). Among Proteobacteria, Alphaproteobacteria were more present in sea ice (25%) compared with the water column (13 and 12% at 5 and 40–60 m, respectively). Inversely, the proportion of Flavobacteriia was higher in the water column (21 and 15% at 2 and 40 m, respectively) when compared with the sea ice (11%). Twenty-nine bacterial species (69%) were specific to a given environment. Only five bacterial isolates (12%) were found in both the sea ice and the water column at both depths, with the closest relatives being Alteromonas stellipolaris (Gammaproteobacteria) [53], Paraglaciecola chatamensis (Gammaproteobacteria) [54,55], Psychrobacter aquimaris (Gammaproteobacteria) [56], Leeuwenhoekiella aequorea (Bacteroidetes) [57] and Rhodococcus cerastii (Actinobacteria) [58].
(b). Growth of Arctic microalgae and characterization of the dissolved organic exudates
The two Arctic diatoms, C. neogracilis and F. cylindrus, were characterized by similar growth rates (0.25 ± 0.02 and 0.30 ± 0.002 d−1, respectively) and reached the stationary phase after 12 and 24 days, respectively. Inorganic nutrient analyses showed that both diatoms cultures were limited mainly by Si(OH)4 in the stationary phase (table 2).
Table 2.
Parameters measured in the microalgae cultures at beginning (T0) and after 44 days (T44d). DOC: dissolved organic carbon; POC: particulate organic carbon; DFCHO-C: dissolved-free monosaccharides; DPCHO-C: dissolved polysaccharides; TDCHO-C: total dissolved saccharides; ND: not determined.
|
C. neogracilis |
F. cylindrus |
|||
|---|---|---|---|---|
| T0 | T44d | T0 | T44d | |
| cells × 104 ml−1 | 8.7 (±2.9) | 122 (±13) | 0.6 (±0.2) | 31.2 (±2.2) |
| NO3 (μM) | 864.9 (±ND) | 224.3 (±118.2) | 859.4 (±ND) | 674.5 (±60.2) |
| PO4 (μM) | 81.5 (±ND) | 20.7 (±8.7) | 79.9 (±ND) | 62.1 (±0.8) |
| Si(OH)4 (μM) | 35.2 (±ND) | 3.6 (±0.7) | 36.0 (±ND) | 2.7 (±1.2) |
| DOC (μM) | 234.1 (±0.8) | 2007.2 (±778.4) | 108.8 (±6.8) | 977 (±70.2) |
| POC (μM) | ND | 2500 (±423) | ND | 612 (±ND) |
| DOC/(POC + DOC) (%) | ND | 45 | ND | 61 |
| DFCHO-C (μM) | ND | 118.2 | ND | 20.2 |
| PCHO-C (μM) | ND | 246.4 | ND | 14.6 |
| TCHO-C (μM) | ND | 364.6 | ND | 34.8 |
| TCHO-C/DOC (%) | ND | 18 | ND | 4 |
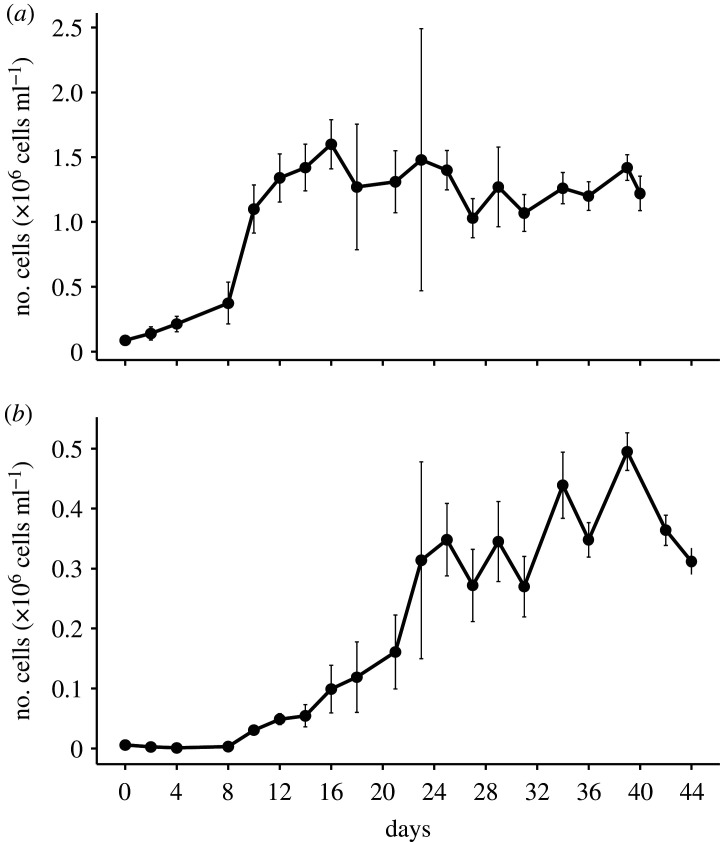
The exudates were recovered from the cultures after 40 days of cultivation for C. neogracilis and after 44 days for F. cylindrus, corresponding to a stationary phase of 32 days for C. neogracilis and 20 days for F. cylindrus (figure 3). In terms of diatom cell abundance, the stationary phase was stable for each strain until this date, suggesting that cell lysis and the consequent release of DOM by this process were of minor importance. DOC concentrations were 234 and 109 µM for C. neogracilis and F. cylindrus, respectively, at the start of the cultures, and they were significantly higher at the end of the experiment due to DOM exudation (table 2). This exudation (DOCfinal − DOCinitial) represented ≈1800 µM for C. neogracilis and ≈870 µM for F. cylindrus. The DOC exuded represented 45% of the sum of POC and DOC for C. neogracilis and 61% for F. cylindrus. Carbohydrates accounted for 4–18% of the DOC and were dominated by polysaccharides in the C. neogracilis exudates (table 2).
Figure 3.
Growth of the microalgae (a) C. neogracilis and (b) F. cylindrus. Values stand for means of triplicate cultures and error bars show ±s.d.
The diatom cultures were not axenic, and an increase in heterotrophic bacterial abundance over time was observed in the cultures (electronic supplementary material, figure S3). We estimated the bacterial consumption of DOC considering a carbon conversion factor of 20 fg C cell−1 [59] and a bacterial growth efficiency of 50% for bacteria growing under high primary productivity conditions [60]. Integrated over the 40–44 days, heterotrophic bacteria accounted for the processing of 0.01% (0.26 µM DOC) and 6% (56 µM DOC) of the DOC produced by C. neogracilis and F. cylindrus, respectively. These results indicate that DOM present in the diatom cultures after 44 days can be considered predominantly diatom-derived and weakly reshuffled by bacteria.
(c). Bioavailability of dissolved organic exudates for the different bacterial strains
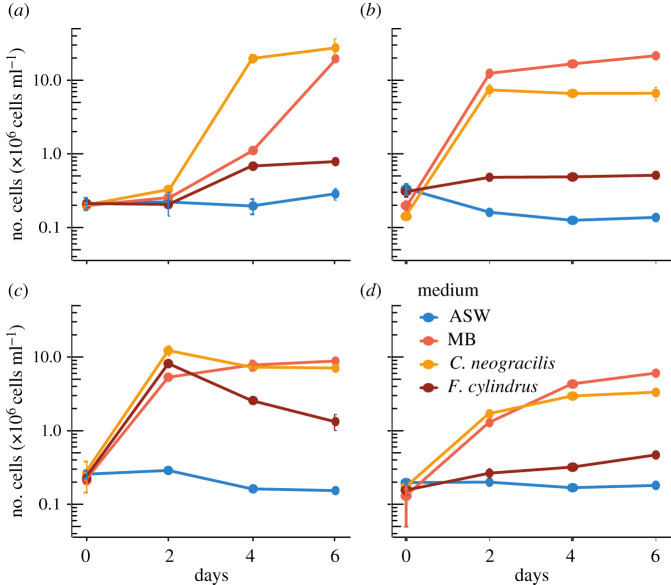
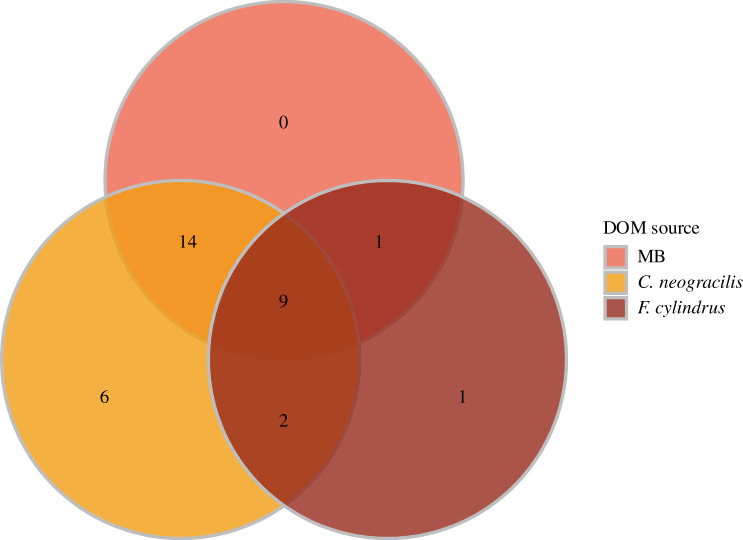
We observed different growth patterns among the strains, indicative of their capacity to use the exudates (figure 4). According to the first index, we confirmed the lack of growth in the negative controls (Artificial Sea Water (ASW) without DOM addition). Surprisingly, only 60% of the strains were able to grow in the diluted MB used as a positive control. When the exudates were used as growth substrates, 78% of bacterial strains (31 out of 40) responded positively to the DOM from C. neogracilis and 33% to the F. cylindrus exudates. The Venn diagram based on these growth capacities shows that only nine strains (23%) were able to grow in all different media (i.e. diluted MB and the two exudates) (figure 5). These strains belonged to the phyla Proteobacteria, Bacteroidetes and Actinobacteria. If we focus on the use of exudates, we found that 55% of the strains were able to use only a single exudate and 27.5% grew on the two exudates. Finally, 17.5% of the strains tested did not grow in any of the two exudates with the majority of bacteria belonging to the Gammaproteobacteria class. We did not find specific relationships between the habitat of the bacterial strains (sea ice bottom versus water column) and their capacity to use the different exudates. For instance, among the 13 bacterial strains able to grow on the exudate of the ice diatom F. cylindrus, only three were isolated from the sea ice. Venn diagrams for each type of habitat are available in electronic supplementary material, figure S4.
Figure 4.
Growth of (a) strain BBCC 770, Polaribacter filamentus (3 m depth, Beaufort Sea); (b) strain BBCC 2586, Idiomarina abyssalis (40 m depth, Green Edge ice camp); (c) strain BBCC 2589, Mesonia algae (40 m depth, Green Edge ice camp); (d) strain BBCC 2637, Stenotrophomonas chelatiphaga (sea ice, Green Edge ice camp); with different sources of DOM: absence of DOM (ASW); diluted MB; exudates from C. neogracilis and F. cylindrus. Values stand for means of triplicate treatments and error bars show ±s.d. (Online version in colour.)
Figure 5.
Venn diagram showing the number of bacterial strains growing with the different microalgae exudates (C. neogracilis, F. cylindrus) and diluted MB. (Online version in colour.)
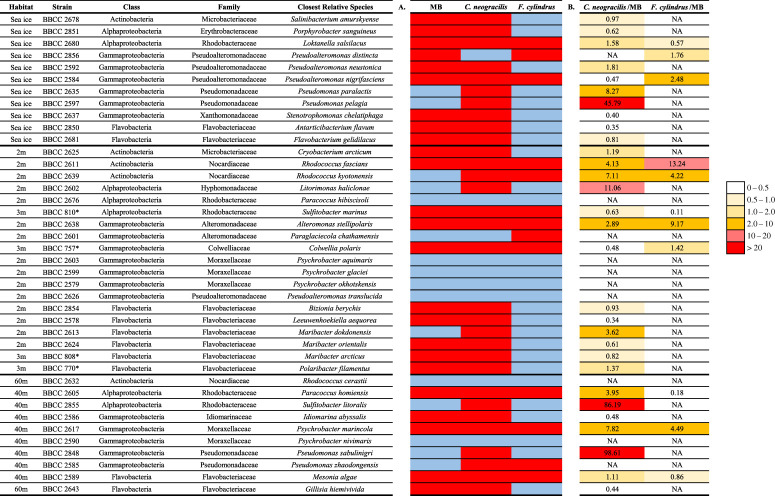
Several isolates belonging to the same genus, such as Rhodococcus, Maribacter and Pseudoalteromonas, had different growth patterns on the same type of exudate (table 3). All Flavobacteriia strains succeeded in degrading the exudate of C. neogracilis. One-third of the bacterial strains obtained an index higher than two using the exudate of C. neogracilis, while this was observed for only 13% of the strains for the F. cylindrus exudate. Index 2 highlights the large variability in the amplitude of growth on exudates relative to a common reference (diluted MB). The highest values (up to 100 times higher than in diluted MB medium) were measured with C. neogracilis exudates during the growth of two Pseudomonas sp. strains and one Sulfitobacter strain (table 2).
Table 3.
Indices characterizing the growth of bacterial strains according to the substrate added. (a) Index 1: distinction between strains which have shown significant growth using different substrates (in red) and those which have not shown visible growth (in blue). (b) Index 2: results of cell growth on different exudates expressed in relation to those measured in diluted MB. See the Material and methods for the explanations of the calculation of the indexes. *Bacterial strains isolated in 2009 in the Beaufort Sea, Arctic Ocean.
 |
4. Discussion
(a). Cultivable bacterial diversity
Isolation of bacterial strains by culturing techniques is key for studying their physiology and functional capabilities. The environmental relevance of isolated strains remains, however, an important issue. Cultivation on a medium rich in organic matter leads to a well-known bias by selecting copiotrophic species that are not necessarily abundant in the marine environment, except during episodes of higher productivity or significant organic matter input [61–63]. Gammaproteobacteria are generally over-represented in the cultivable fraction compared with the natural community [64]. In our case, they represented 53% of the isolates, whereas in situ bacterial community composition determined by Illumina sequencing showed that the relative abundance of Gammaproteobacteria was 47% in sea ice and 31% in the water column [33]. Strains belonging to the Flavobacteriaceae family were well represented among the isolated strains (n = 16; 17% of the total strains isolated). Interestingly, the survey of the bacterial community composition by metabarcoding underlined the importance of this family during the phytoplankton bloom in this region [33]. Indeed, Flavobacteriaceae were well represented in the total in situ bacterial community with 16% of the total DNA sequences. Furthermore, they reflected 19% of the sequences in the sea ice bottom and 16% of the sequences in the water column. The Rhodobaceteraceae bacterial family was well represented in the isolates with 10 bacterial strains. As with Flavobacteriaceae, Rhodobaceteraceae members are frequently associated with phytoplankton blooms in marine ecosystems [20].
In the sea ice bottom, the cultivable genera belonging to Paraglaciecola (Gammaproteobacteria) represented 12% of the sequences obtained by Illumina sequencing from the in situ community at the same period, Polaribacter (Flavobacteriia) represented 8%, Pseudoalteromonas (Gammaproteobacteria) represented 6% and Colwellia (Gammaproteobacteria) 1% [33]. In the water column, the cultivable genera with the highest in situ abundance were Polaribacter (Flavobacteriia) with 7%, Pseudoalteromonas (Gammaproteobacteria) with 3%, Paraglaciecola (Gammaproteobacteria) with 2%, Rhodococcus (Actinobacetria) with 2% and Pseudomonas (Gammaproteobacteria) with 1% of total sequences. This is a rough estimate, because each genus may contain many different species, of which only a few representatives have been isolated. However, these results underline the fact that the isolated strains were overall representative of the in situ bacterial community, especially in the sea ice bottom.
Finally, comparison of the sequences of our isolated strains with sequences in the EZBiocloud database revealed that the lowest percentage of similarity (95.72%) was obtained for Antarcticibacterium flavum [65] belonging to the class of Flavobacteriia (Bacteroidetes). This low percentage of similarity suggests the isolation of a new species belonging to the genus Antarcticibacterium which can only be confirmed by further genetic analysis.
(b). Biodegradability of the microalgae exudates
To assess how phytoplanktonic blooms can shape bacterial community composition, we performed a biodegradation assay to identify the capacity of several isolated bacterial strains to use the DOM produced by two polar diatoms, C. neogracilis and F. cylindrus. The exudates used in the present study originated from non-axenic diatom cultures. We estimated that heterotrophic bacteria consumed up to 6% of the DOC over the course of culturing period, and a fraction of this organic carbon could have been released by bacteria as DOM with a chemical composition different from the consumed compounds [66,67]. Therefore, part of the readily available substrates specific for each diatom species was likely used or transformed prior to the biodegradation experiments. Given the overall low fraction of bacterially processed organic carbon and exudation being a continuous process throughout the culturing period, we consider the DOM used here representative of the two diatoms. This idea is further supported by our biodegradation assays that revealed a preference of the bacterial strains for the use of DOM from C. neogracilis, the diatom with the longer stationary phase, when compared with F. cylindrus.
We investigated carbohydrates, major components of phytoplankton exudates, to explore differences in the chemical composition of the two DOM sources. The relative abundance of carbohydrates in phytoplankton exudates appears variable among phytoplankton species (26–80% [68]; 6–13% [28]) and tends to increase in the stationary phase [68]. These two facts can explain why F. cylindrus exudates had a lower contribution of carbohydrates to the DOM (4%) when compared with C. neogracilis (18%). Polymeric and monomeric carbohydrates were consistently shown to be used in short-term (days) biodegradation experiments [69,70]. The lower content of this labile source of organic carbon in the exudate of the sea ice diatom when compared with the water column diatom could explain the lower number of bacterial strains growing on this DOM. Carbohydrates are present in high concentrations (80–1300 µMC) in sea ice when compared with the water column in the Arctic Ocean [71,72]. The high similarity observed in other studies between the carbohydrate composition of sea ice and the carbohydrates produced by sea ice diatoms, including F. cylindrus, confirm the microalgal origin of these compounds [72].
Furthermore, we did not find a specific relationship between the habitat of bacterial strains and their capacity to use the different exudates. As explained earlier, among the 13 bacterial strains able to grow on the F. cylindrus exudate, only three strains have been isolated from sea ice samples. This may indicate that a co-metabolism is important to degrade such exudates in the sea ice environment. Indeed, some bacterial strains could be able to degrade the primary energy source given by the F. cylindrus DOM in the ice, producing secondary compounds which in turn would be used by other bacterial strains as potentially the ones which did not grow in our biodegradation assay. Thus, our results could complement previous observations with mixed communities where cross-feeding among taxa can occur [73]. However, six strains out of 13 (46%) which grew on this DOM were isolated at 2 and 3 m depth. Thus, it means that 69% of the strains growing on F. cylindrus DOM were isolated in the ice or in the first metres of the water column. This is relevant to the fact that some interactions happen between the sea ice bottom and the water column. The microalgae living in the ice excrete DOM will be used as a source of carbon by bacteria in the first few metres of the water column. Moreover, these interactions are reinforced as spring progresses with the beginning of the sea ice melt causing the release of a significant amount of DOM into the water column, which will serve as a source of energy and carbon for bacteria [33].
The growth of bacterial strains on the diatom exudates is in part linked to their capacity to produce extracellular enzymes to degrade polymers. In the present study, polysaccharides constituted a lower fraction of the carbohydrates of the F. cylindrus exudate (42%) when compared with the C. neogracilis exudate (68%) (table 2). The incubation of phylogenetically different bacterial strains with a given DOM source allowed us to determine that the genetic potential to produce extracellular enzymes, more or less specific to DOM compounds, was a selective mechanism for the use of a given exudate. Phytoplankton-derived polysaccharide composition is highly diverse, may fluctuate between species [74] and is influenced by external factors such as nutrients, light and salinity stress [75]. Jain & Krishnan [76] showed that 52% of a total of 215 cold-adapted heterotrophic bacteria (all belonging to Gammaproteobacteria) isolated in an open glacial fjord in the Arctic Ocean were able to produce extracellular polysaccharide enzymes at 4°C. Interestingly, these authors observed high diversity in terms of enzyme activities among strains belonging to a same genus and/or species. This result can be compared with the observation we made in the heterogeneity of the growth response within the same genus for the same type of exudate. Whether a DOM source is biologically labile might therefore depend on the bacterial taxa and their respective functional repertoire.
The commonly observed rapid utilization of phytoplankton exudates by mixed microbial communities is likely due to the activity of the most successful taxa. In this context, it was interesting to note that the highest growth was observed for one Sulfitobacter strain on the C. neogracilis exudate. Two Sulfitobacter Operational Taxonomic Units (OTUs) were identified as most responsive among a Southern Ocean natural bacterial community to the exudate of Chaetoceros debilis [77]. This bacterial group is known for its capacity to use organic sulfur compounds that could be the basis for this distinct diatom–bacteria association. Sulfitobacter could have responded positively to the dimethylsulfide produced in the C. neogracilis culture, even if this diatom has been shown to emit this compound at a lower rate when compared with Emiliania huxleyi [78].
All Flavobacteriia, including a representative of the genus Polaribacter, had the capacity to use the C. neogracilis exudate. Flavobacteriia and more precisely the genus Polaribacter were identified as a bacterial group responding positively to diatom blooms in polar regions [30,33]. Recently, we confirmed the responsiveness of Polaribacter after the addition of C. neogracilis exudates to a natural bacterial community present in the Baffin Bay at two ice zone stations [31]. Marine Flavobacteriia are known as efficient degraders of biopolymers, such as proteins and polysaccharides [79]. Over the last decade, next-generation sequencing technologies have enabled the emergence of a detailed analysis of the response of several cultivable bacterial strains, representative of Flavobacteriia, to organic matter, in particular through proteomic and transcriptomic approaches [80]. This approach was used to explore the metabolic pathways of two Polaribacter strains isolated in the North Sea during their growth with various models of organic molecules like carbohydrates and polysaccharides [80]. This study showed that both Polaribacter strains were able to degrade algal polysaccharides, while they each occupy different ecologic niches. However, they observed that one strain was feeding on proteins and a small subset of algal polysaccharides, while the other was able to degrade a wider range of polysaccharides, making it a more flexible strain [80]. More recently, proteomic approaches have also been used to explore the response of Formosa strains (Flavobacteriaceae) isolated in the North Sea to laminarin as a model of polysaccharides [81]. The authors observed that laminarin was used as a major source of energy, and as a signal molecule, which induced transporters and digestive enzymes to also use other compounds released from the lysis of diatom cells. These studies confirm the link between Flavobacteria and microalgal blooms with their capacity to degrade and use algal DOM. It is consistent with the fact that all the bacterial strains belonging to Flavobacteria tested in our biodegradation assay were able to grow on the pelagic microalgae C. neogracilis exudate. However, depending on their genetic material, bacterial strains differ in their ability to degrade DOM according to its composition. It implies the occurrence of different ecological niches in which bacterial strains use the energy source to which they are the most adapted.
Even though the environmental relevance of isolated strains remains an important issue, our study underlines the fact that the isolated strains from the ice camp were overall representative of the in situ bacterial community, especially in the sea ice bottom. We showed through a biodegradation experiment a preference for the DOM degradation from the pelagic diatom C. neogracilis among 40 different bacterial strains isolated from ice and several depths. Indeed, most of the strains tested (78%) were able to grow on the exudate from the pelagic diatom C. neogracilis, and 33% were able to use the exudate from the sea ice diatom F. cylindrus. 17.5% of the strains were not able to grow with any exudate, while 27.5% of the strains were able to use both types of exudates. Nevertheless, we observed highly diverse response patterns among the bacterial strains even between strains with the same genus. We did not find specific relationships between the habitat of the bacterial strains (sea ice bottom versus water column) and their capacity to use the different exudates. One hypothesis for this lack of relationship between microbial habitat and microalgal DOM could be cross-feeding, meaning the production of secondary compounds based on the degradation of microalgal DOM by specific bacterial strains which allows less specific strains to use these secondary compounds for their growth. All strains belonging to Flavobacteriia (n = 10) were able to use the DOM provided by C. neogracilis, and this exudate sustained a growth capacity of up to 100 times higher than diluted MB medium, of two Pseudomonas sp. strains and one Sulfitobacter strain.
In conclusion, using a simple model of interaction (one microalgal exudate/one bacterial strain) to avoid complex biotic interactions which occur in microbial communities, we were able to confirm that dissolved exudates from different microalgae differed in their bioavailability to bacterial strains. Our results reinforce the fact that DOM can shape the bacterial community composition by selecting the most responsive species having the right genetic material to degrade the DOM at their disposal in their specific environment.
Supplementary Material
Acknowledgements
We thank the Roscoff Culture Collection (RCC) for proving us the microalgae strains. We thank J. Caparros for DOC and POC analyses, O. Crispi for inorganic nutrient analyses, D. Vaulot, D. Marie and M. Tragin for bacterial counts by flow cytometry, J. Ras and C. Dimier for chlorophyll a measurements by HPLC. We are grateful to the BIO2MAR and the BIOPIC platforms (http://bio2mar.obs-banyuls.fr/) for providing technical support and access to instrumentation. The field campaign would not have been possible without the support of the Hamlet of Qikiqtarjuaq and the members of the community as well as the Inuksuit School and its Principal, Jacqueline Arsenault. The field campaign was successful thanks to the contribution of J. Ferland, G. Bécu, C. Marec, J. Lagunas, F. Bruyant, J. Larivière, E. Rehm, S. Lambert-Girard, C. Aubry, C. Lalande, A. LeBaron, C. Marty, J. Sansoulet, D. Christiansen-Stowe, A. Wells, M. Benoît-Gagné, E. Devred and M.-H. Forget from the Takuvik laboratory, C.J. Mundy and V. Galindo from the University of Manitoba & F. Pinczon du Sel and E. Brossier from Vagabond. We also thank Michel Gosselin, Québec-Océan, the CCGS Amundsen and the Polar Continental Shelf Program for their in-kind contribution in terms of polar logistics and scientific equipment.
Data accessibility
All DNA sequences were deposited in GenBank (NCBI) with accession nos. MK 224724–224815 for bacterial strains isolated during this study and MK 217878–217881 for four bacterial strains isolated in the Beaufort Sea. Additional data are available within the electronic supplementary material.
Authors' contributions
F.J. and L.T. conceived and designed the research, L.T., L.D., L.I., P.C. and C.P. performed the experimental work, L.T., L.D., L.I., P.C., C.P., I.O. and F.J. contributed to the interpretation of the data and the discussion of the results presented in the manuscript. F.J. wrote the first draft of the manuscript and all the authors made comments and amendments, and approved the final version.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by LEFE-CYBER CNRS program (EXPERT project) and by the following French and Canadian programs and agencies (Green Edge project): ANR (contract no. 111112), ArcticNet, CERC on Remote sensing of Canada's new Arctic frontier, CNES (project no. 131425), French Arctic Initiative, Fondation Total, CSA, LEFE and IPEV (project no. 1164). This latest project was conducted under the scientific coordination of the Canada Excellence Research Chair on Remote sensing of Canada's new Arctic frontier and the CNRS & Université Laval Takuvik Joint International Laboratory (UMI3376). L.D. and L.T. were supported by PhD grants from Sorbonne Université.
References
- 1.Nagata T. 2000. Production mechanisms of dissolved organic matter. In Microbial ecology of the oceans (ed. Kirchman DL.), pp. 121–152. New York: Wiley-Liss. [Google Scholar]
- 2.Thornton DCO. 2014. Dissolved organic matter (DOM) release by phytoplankton in the contemporary and future ocean. Eur. J. Phycol. 49, 20–46. ( 10.1080/09670262.2013.875596) [DOI] [Google Scholar]
- 3.Engel A, Borchard C, Piontek J, Schulz KG, Riebesell U, Bellerby R. 2013. CO2 increases 14C primary production in an Arctic plankton community. Biogeosciences 10, 1291–1308. ( 10.5194/bg-10-1291-2013) [DOI] [Google Scholar]
- 4.Obernosterer I, Herndl GJ. 1995. Phytoplankton extracellular release and bacterial growth: dependence on the inorganic N:P ratio. Mar. Ecol. Prog. Ser. 116, 247–257. ( 10.3354/meps116247) [DOI] [Google Scholar]
- 5.Box JE, et al. 2019. Key indicators of Arctic climate change: 1971–2017. Environ. Res. Lett. 14, 045010 ( 10.1088/1748-9326/aafc1b) [DOI] [Google Scholar]
- 6.Randelhoff A, et al. 2019. The evolution of light and vertical mixing across a phytoplankton ice-edge bloom. Elem. Sci. Anthr. 7, 20 ( 10.1525/elementa.357) [DOI] [Google Scholar]
- 7.Wassmann P, Reigstad M. 2011. Future Arctic Ocean seasonal ice zones and implications for pelagic-benthic coupling. Oceanography 24, 220–231. ( 10.5670/oceanog.2011.65) [DOI] [Google Scholar]
- 8.Arrigo KR, van Dijken GL.. 2011. Secular trends in Arctic Ocean net primary production. J. Geophys. Res. Ocean. 116, 1–15. ( 10.1029/2011JC007151) [DOI] [Google Scholar]
- 9.Pabi S, van Dijken GL, Arrigo KR. 2008. Primary production in the Arctic Ocean, 1998–2006. J. Geophys. Res. Ocean. 113, 1998–2006. ( 10.1029/2007JC004578) [DOI] [Google Scholar]
- 10.Arrigo KR, et al. 2012. Massive phytoplankton blooms under arctic sea ice. Science 336, 1408 ( 10.1126/science.1215065) [DOI] [PubMed] [Google Scholar]
- 11.Arrigo KR, et al. 2014. Phytoplankton blooms beneath the sea ice in the Chukchi Sea. Deep Res. Part II Top. Stud. Oceanogr. 105, 1–16. ( 10.1016/j.dsr2.2014.03.018) [DOI] [Google Scholar]
- 12.Lowry KE, van Dijken GL, Arrigo KR.. 2014. Evidence of under-ice phytoplankton blooms in the Chukchi Sea from 1998 to 2012. Deep Res. Part II Top. Stud. Oceanogr. 105, 105–117. ( 10.1016/j.dsr2.2014.03.013) [DOI] [Google Scholar]
- 13.Assmy P, et al. 2017. Leads in Arctic pack ice enable early phytoplankton blooms below snow-covered sea ice. Sci. Rep. 7, 1–9. ( 10.1038/srep40850) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Quillfeldt CH. 2000. Common diatom species in Arctic spring blooms: their distribution and abundance. Bot. Mar. 43, 499–516. ( 10.1515/BOT.2000.050) [DOI] [Google Scholar]
- 15.Laney SR, Sosik HM. 2014. Phytoplankton assemblage structure in and around a massive under-ice bloom in the Chukchi Sea. Deep Res. Part II Top. Stud. Oceanogr. 105, 30–41. ( 10.1016/j.dsr2.2014.03.012) [DOI] [Google Scholar]
- 16.Leu E, Mundy CJ, Assmy P, Campbell K, Gabrielsen TM, Gosselin M, Juul-Pedersen T, Gradinger R. 2015. Arctic spring awakening—Steering principles behind the phenology of vernal ice algal blooms. Prog. Oceanogr. 139, 151–170. ( 10.1016/j.pocean.2015.07.012) [DOI] [Google Scholar]
- 17.van Leeuwe MA, et al. 2018. Microalgal community structure and primary production in Arctic and Antarctic sea ice: a synthesis. Elem. Sci. Anth. 6, 4 ( 10.1525/elementa.267) [DOI] [Google Scholar]
- 18.Cotner JB, Biddanda BA. 2002. Small players, large role: microbial influence on biogeochemical processes in pelagic aquatic ecosystems. Ecosystems 5, 105–121. ( 10.1007/s10021-001-0059-3) [DOI] [Google Scholar]
- 19.Azam F, Fenchel T, Field J, Gray J, Meyer-Reil L, Thingstad F. 1983. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263. ( 10.3354/meps010257) [DOI] [Google Scholar]
- 20.Buchan A, LeCleir GR, Gulvik CA, González JM. 2014. Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698. ( 10.1038/nrmicro3326) [DOI] [PubMed] [Google Scholar]
- 21.Amin SA, Parker MS, Armbrust EV. 2012. Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 76, 667–684. ( 10.1128/mmbr.00007-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paul C, Pohnert G. 2011. Interactions of the algicidal bacterium Kordia algicida with diatoms: regulated protease excretion for specific algal lysis. PLoS ONE 6, e21032 ( 10.1371/journal.pone.0021032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durham BP, et al. 2015. Cryptic carbon and sulfur cycling between surface ocean plankton. Proc. Natl. Acad. Sci. USA 112, 453–457. ( 10.1073/pnas.1413137112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gärdes A, Ramaye Y, Grossart HP, Passow U, Ullrich MS. 2012. Effects of Marinobacter adhaerens HP15 on polymer exudation by Thalassiosira weissflogii at different N:P ratios. Mar. Ecol. Prog. Ser. 461, 1–14. ( 10.3354/meps09894) [DOI] [Google Scholar]
- 25.Koedooder C, Stock W, Willems A, Mangelinckx S, De Troch M, Vyverman W, Sabbe K.. 2019. Diatom-bacteria interactions modulate the composition and productivity of benthic diatom biofilms. Front. Microbiol. 10, 1255 ( 10.3389/fmicb.2019.01255) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Underwood GJC, Michel C, Meisterhans G, Niemi A, Belzile C, Witt M, Dumbrell AJ, Koch BP. 2019. Organic matter from Arctic sea-ice loss alters bacterial community structure and function. Nat. Clim. Chang. 9, 170–176. ( 10.1038/s41558-018-0391-7) [DOI] [Google Scholar]
- 27.Tada Y, Nakaya R, Goto S, Yamashita Y, Suzuki K. 2017. Distinct bacterial community and diversity shifts after phytoplankton-derived dissolved organic matter addition in a coastal environment. J. Exp. Mar. Biol. Ecol. 495, 119–128. ( 10.1016/j.jembe.2017.06.006) [DOI] [Google Scholar]
- 28.Landa M, et al. 2014. Phylogenetic and structural response of heterotrophic bacteria to dissolved organic matter of different chemical composition in a continuous culture study. Environ. Microbiol. 16, 1668–1681. ( 10.1111/1462-2920.12242) [DOI] [PubMed] [Google Scholar]
- 29.Sarmento H, Morana C, Gasol JM. 2016. Bacterioplankton niche partitioning in the use of phytoplankton-derived dissolved organic carbon: quantity is more important than quality. ISME J. 10, 2582–2592. ( 10.1038/ismej.2016.66) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luria CM, Amaral-Zettler LA, Ducklow HW, Rich JJ. 2016. Seasonal succession of free-living bacterial communities in coastal waters of the Western Antarctic Peninsula. Front. Microbiol. 7, 1731 ( 10.3389/fmicb.2016.01731) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dadaglio L, Dinasquet J, Obernosterer I, Joux F. 2018. Differential responses of bacteria to diatom-derived dissolved organic matter in the Arctic Ocean. Aquat. Microb. Ecol. 82, 59–72. ( 10.3354/ame01883) [DOI] [Google Scholar]
- 32.Oziel L, et al. 2019. Environmental factors influencing the seasonal dynamics of spring algal blooms in and beneath sea ice in western Baffin Bay. Elem. Sci. Anth. 7, 34 ( 10.1525/elementa.372) [DOI] [Google Scholar]
- 33.Dadaglio L. 2018. Dynamique des communautés bactériennes en réponse au bloom phytoplanctonique dans l'océan Arctique et identification des acteurs microbiens impliqués dans la dégradation de la matière organique. Paris, France: Sorbonne Université. [Google Scholar]
- 34.Balzano S, Marie D, Gourvil P, Vaulot D. 2012. Composition of the summer photosynthetic pico and nanoplankton communities in the Beaufort Sea assessed by T-RFLP and sequences of the 18S rRNA gene from flow cytometry sorted samples. ISME J. 6, 1480–1498. ( 10.1038/ismej.2011.213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aslam SN, Strauss J, Thomas DN, Mock T, Underwood GJC. 2018. Identifying metabolic pathways for production of extracellular polymeric substances by the diatom Fragilariopsis cylindrus inhabiting sea ice. ISME J. 12, 1237–1251. ( 10.1038/s41396-017-0039-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zobell CE. 1941. Studies on marine bacteria. I. The cultural requirements of heterotrophic aerobes. J. Mar. Res. 4, 42–75. [Google Scholar]
- 37.Hogg JC, Lehane MJ. 1999. Identification of bacterial species associated with the sheep scab mite (Psoroptes ovis) by using amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 65, 4227–4229. ( 10.1128/AEM.65.9.4227-4229.1999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. 2008. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74, 2461–2470. ( 10.1128/AEM.02272-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanger F, Nicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74, 5463–5467. ( 10.1073/pnas.74.12.5463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morales SE, Holben WE. 2009. Empirical testing of 16S rRNA gene PCR primer pairs reveals variance in target specificity and efficacy not suggested by in silico analysis. Appl. Environ. Microbiol. 75, 2677–2683. ( 10.1128/AEM.02166-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Staden R, Judge DP, Bonfield JK. 2003. Managing sequencing projects in the GAP4 environment. In Introduction to bioinformatics (eds Krawetz SA, Womble DD), pp. 327–344. Totowa, NJ: Humana Press. [Google Scholar]
- 42.Kim OS, et al. 2012. Introducing EzTaxon-e: a prokaryotic 16 s rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721. ( 10.1099/ijs.0.038075-0) [DOI] [PubMed] [Google Scholar]
- 43.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. ( 10.1093/nar/gkh340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. ( 10.1093/molbev/msy096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.VanLandingham SL. 1968. Catalogue of the fossil and recent genera and species of diatoms and their synonyms; part II, Bacteriastrum through Coscinodiscus. Vaduz, Liechtenstein: See https://eurekamag.com/research/018/519/018519603.php. [Google Scholar]
- 46.Keller MD, Selvin RC, Claus W, Guillard RRL. 1987. Media for the culture of oceanic ultraphytoplankton. J. Phycol. 23, 633–638. ( 10.1111/j.1529-8817.1987.tb04217.x) [DOI] [Google Scholar]
- 47.Guillard RRL, Hargraves PE. 1993. Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia 32, 234–236. ( 10.2216/i0031-8884-32-3-234.1) [DOI] [Google Scholar]
- 48.Myklestad SM, Skånøy E, Hestmann S. 1997. A sensitive and rapid method for analysis of dissolved mono- and polysaccharides in seawater. Mar. Chem. 56, 279–286. ( 10.1016/S0304-4203(96)00074-6) [DOI] [Google Scholar]
- 49.Panagiotopoulos C, Sempéré R. 2005. Analytical methods for the determination of sugars in marine samples: a historical perspective and future directions. Limnol. Oceanogr. Methods 3, 419–454. ( 10.4319/lom.2005.3.419) [DOI] [Google Scholar]
- 50.Panagiotopoulos C, Sempéré R, Jacq V, Charrière B. 2014. Composition and distribution of dissolved carbohydrates in the Beaufort Sea Mackenzie margin (Arctic Ocean). Mar. Chem. 166, 92–102. ( 10.1016/j.marchem.2014.09.004) [DOI] [Google Scholar]
- 51.Ortega-Retuerta E, Joux F, Jeffrey WH, Ghiglione JF. 2013. Spatial variability of particle-attached and free-living bacterial diversity in surface waters from the Mackenzie River to the Beaufort Sea (Canadian Arctic). Biogeosciences 10, 2747–2759. ( 10.5194/bg-10-2747-2013) [DOI] [Google Scholar]
- 52.Eguchi M, Nishikawa T, McDonald K, Cavicchioli R, Gottschal JC, Kjelleberg S. 1996. Responses to stress and nutrient availability by the marine ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 62, 1287–1294. ( 10.1128/AEM.62.4.1287-1294.1996) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Trappen S, Tan TL, Yang J, Mergaert J, Swings J.. 2004. Alteromonas stellipolaris sp. nov., a novel, budding, prosthecate bacterium from Antarctic seas, and emended description of the genus Alteromonas. Int. J. Syst. Evol. Microbiol. 54, 1157–1163. ( 10.1099/ijs.0.02862-0) [DOI] [PubMed] [Google Scholar]
- 54.Matsuyama H, Hirabayashi T, Kasahara H, Minami H, Hoshino T, Yumoto I. 2006. Glaciecola chathamensis sp. nov., a novel marine polysaccharide-producing bacterium. Int. J. Syst. Evol. Microbiol. 56, 2883–2886. ( 10.1099/ijs.0.64413-0) [DOI] [PubMed] [Google Scholar]
- 55.Shivaji S, Reddy GS. 2014. Phylogenetic analyses of the genus Glaciecola: emended description of the genus Glaciecola, transfer of Glaciecola mesophila, G. agarilytica, G. aquimarina, G. arctica, G. chathamensis, G. polaris and G. psychrophila to the genus Paraglaciecola gen. nov. Int. J. Syst. Evol. Microbiol. 64, 3264–3275. ( 10.1099/ijs.0.065409-0) [DOI] [PubMed] [Google Scholar]
- 56.Yoon JH, Lee CH, Yeo SH, Oh TK. 2005. Psychrobacter aquimaris sp. nov. and Psychrobacter namhaensis sp. nov., isolated from sea water of the South Sea in Korea. Int. J. Syst. Evol. Microbiol. 55, 1007–1013. ( 10.1099/ijs.0.63464-0) [DOI] [PubMed] [Google Scholar]
- 57.Nedashkovskaya OI, et al. 2005. Reclassification of [Cytophaga] marinoflava Reichenbach 1989 as Leeuwenhoekiella marinoflava gen. nov., comb. nov. and description of Leeuwenhoekiella aequorea sp. nov. Int. J. Syst. Evol. Microbiol. 55, 1033–1038. ( 10.1099/ijs.0.63410-0) [DOI] [PubMed] [Google Scholar]
- 58.Kämpfer P, Wellner S, Lohse K, Lodders N, Martin K. 2013. Rhodococcus cerastii sp. nov. and Rhodococcus trifolii sp. nov., two novel species isolated from leaf surfaces. Int. J. Syst. Evol. Microbiol. 63, 1024–1029. ( 10.1099/ijs.0.044958-0) [DOI] [PubMed] [Google Scholar]
- 59.Lee S, Fuhrman JA. 1987. Relationships between biovolume and biomass of naturally derived marine bacterioplankton. Appl. Environ. Microbiol. 53, 1298–1303. ( 10.1128/AEM.53.6.1298-1303.1987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Biddanda B, Opsahl S, Benner R. 1994. Plankton respiration and carbon flux through bacterioplankton on the Louisiana shelf. Limnol. Oceanogr. 39, 1259–1275. ( 10.4319/lo.1994.39.6.1259) [DOI] [Google Scholar]
- 61.Nelson CE, Carlson CA. 2012. Tracking differential incorporation of dissolved organic carbon types among diverse lineages of Sargasso Sea bacterioplankton. Environ. Microbiol. 14, 1500–1516. ( 10.1111/j.1462-2920.2012.02738.x) [DOI] [PubMed] [Google Scholar]
- 62.Blanchet M, et al. 2015. Changes in bacterial community metabolism and composition during the degradation of dissolved organic matter from the jellyfish Aurelia aurita in a Mediterranean coastal lagoon. Environ. Sci. Pollut. Res. 22, 13 638–13 653. ( 10.1007/s11356-014-3848-x) [DOI] [PubMed] [Google Scholar]
- 63.Mayali X, Stewart B, Mabery S, Weber PK. 2016. Temporal succession in carbon incorporation from macromolecules by particle-attached bacteria in marine microcosms. Environ. Microbiol. Rep. 8, 68–75. ( 10.1111/1758-2229.12352) [DOI] [PubMed] [Google Scholar]
- 64.Eilers H, Pernthaler J, Glöckner FO, Amann R. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66, 3044–3051. ( 10.1128/AEM.66.7.3044-3051.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li A, Lin L, Zhang M, Zhu H. 2018. Antarcticibacterium flavum gen. nov., sp. nov., isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 68, 254–259. ( 10.1099/ijsem.0.002489) [DOI] [PubMed] [Google Scholar]
- 66.Ogawa H, Amagai Y, Koike I, Kaiser K, Benner R. 2001. Production of refractory dissolved organic matter by bacteria. Science 292, 917–920. ( 10.1126/science.1057627) [DOI] [PubMed] [Google Scholar]
- 67.Lechtenfeld OJ, Hertkorn N, Shen Y, Witt M, Benner R. 2015. Marine sequestration of carbon in bacterial metabolites. Nat. Commun. 6, 6711 ( 10.1038/ncomms7711) [DOI] [PubMed] [Google Scholar]
- 68.Biddanda B, Benner R. 1997. Carbon, nitrogen, and carbohydrate fluxes during the production of particulate and dissolved organic matter by marine phytoplankton. Limnol. Oceanogr. 42, 506–518. ( 10.4319/lo.1997.42.3.0506) [DOI] [Google Scholar]
- 69.Burney CM. 1986. Bacterial utilization of total in situ dissolved carbohydrate in offshore waters. Limnol. Oceanogr. 31, 427–431. ( 10.4319/lo.1986.31.2.0427) [DOI] [Google Scholar]
- 70.Arnosti C. 2000. Substrate specificity in polysaccharide hydrolysis: contrasts between bottom water and sediments. Limnol. Oceanogr. 45, 1112–1119. ( 10.4319/lo.2000.45.5.1112) [DOI] [Google Scholar]
- 71.Underwood GJC, Aslam SN, Michel C, Niemi A, Norman L, Meiners KM, Laybourn-Parry J, Paterson H, Thoma DN. 2013. Broad-scale predictability of carbohydrates and exopolymers in Antarctic and Arctic sea ice. Proc. Natl. Acad. Sci. USA 110, 15 734–15 739. ( 10.1073/pnas.1302870110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aslam SN, Michel C, Niemi A, Underwood GJC. 2016. Patterns and drivers of carbohydrate budgets in ice algal assemblages from first year Arctic sea ice. Limnol. Oceanogr. 61, 919–937. ( 10.1002/lno.10260) [DOI] [Google Scholar]
- 73.Beier S, Bertilsson S. 2011. Uncoupling of chitinase activity and uptake of hydrolysis products in freshwater bacterioplankton. Limnol. Oceanogr. 56, 1179–1188. ( 10.4319/lo.2011.56.4.1179) [DOI] [Google Scholar]
- 74.Mühlenbruch M, Grossart HP, Eigemann F, Voss M. 2018. Mini-review: phytoplankton-derived polysaccharides in the marine environment and their interactions with heterotrophic bacteria. Environ. Microbiol. 20, 2671–2685. ( 10.1111/1462-2920.14302) [DOI] [PubMed] [Google Scholar]
- 75.Abdullahi AS, Underwood GJC, Gretz MR. 2006. Extracellular matrix assembly in diatoms (Bacillariophyceae). V. Environmental effects on polysaccharide synthesis in the model diatom, Phaeodactylum tricornutum. J. Phycol. 42, 363–378. ( 10.1111/j.1529-8817.2006.00193.x) [DOI] [Google Scholar]
- 76.Jain A, Krishnan KP. 2017. A glimpse of the diversity of complex polysaccharide-degrading culturable bacteria from Kongsfjorden, Arctic Ocean. Ann. Microbiol. 67, 203–214. ( 10.1007/s13213-016-1252-0) [DOI] [Google Scholar]
- 77.Landa M, Blain S, Harmand J, Monchy S, Rapaport A, Obernosterer I. 2018. Major changes in the composition of a Southern Ocean bacterial community in response to diatom-derived dissolved organic matter. FEMS Microbiol. Ecol. 94, 34 ( 10.1093/femsec/fiy034) [DOI] [PubMed] [Google Scholar]
- 78.Yassaa N, Colomb A, Lochte K, Peeken I, Williams J. 2006. Development and application of a headspace solid-phase microextraction and gas chromatography/mass spectrometry method for the determination of dimethylsulfide emitted by eight marine phytoplankton species. Limnol. Oceanogr. Methods 4, 374–381. ( 10.4319/lom.2006.4.374) [DOI] [Google Scholar]
- 79.Williams TJ, Wilkins D, Long E, Evans F, DeMaere MZ, Raftery MJ, Cavicchioli R. 2013. The role of planktonic Flavobacteria in processing algal organic matter in coastal East Antarctica revealed using metagenomics and metaproteomics. Environ. Microbiol. 15, 1302–1317. ( 10.1111/1462-2920.12017) [DOI] [PubMed] [Google Scholar]
- 80.Xing P, et al. 2015. Niches of two polysaccharide-degrading Polaribacter isolates from the North Sea during a spring diatom bloom. ISME J. 9, 1410–1422. ( 10.1038/ismej.2014.225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Unfried F, et al. 2018. Adaptive mechanisms that provide competitive advantages to marine bacteroidetes during microalgal blooms. ISME J. 12, 2894–2906. ( 10.1038/s41396-018-0243-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All DNA sequences were deposited in GenBank (NCBI) with accession nos. MK 224724–224815 for bacterial strains isolated during this study and MK 217878–217881 for four bacterial strains isolated in the Beaufort Sea. Additional data are available within the electronic supplementary material.