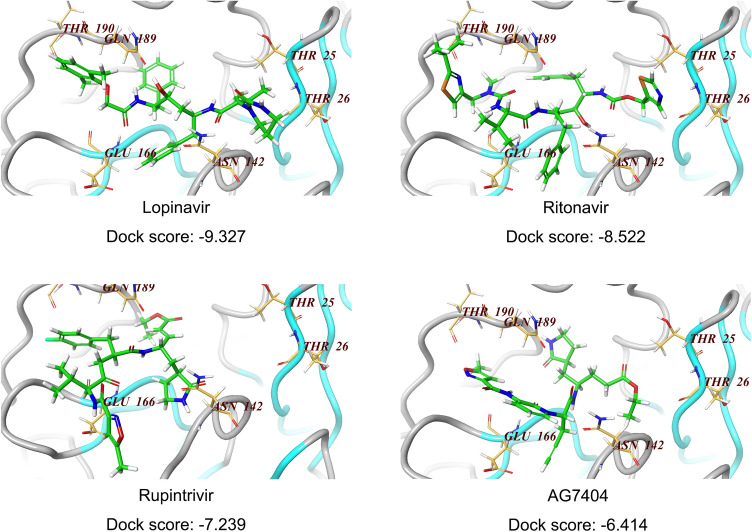
Fig. 4.
Docking of compounds on active site of SARS-CoV-2 3CLpro homology model. For molecular docking, the compounds (lopinavir, ritonavir, rupintrivir, AG7404) were built and subjected to the optimize geometry calculation in mechanics using MMFF94 parameter at DS2.5. The compounds were then docked at the homology model of SARS-CoV-2 3CLpro using Schrodinger Glide program. The docking scores of these four compounds were calculated and listed below.