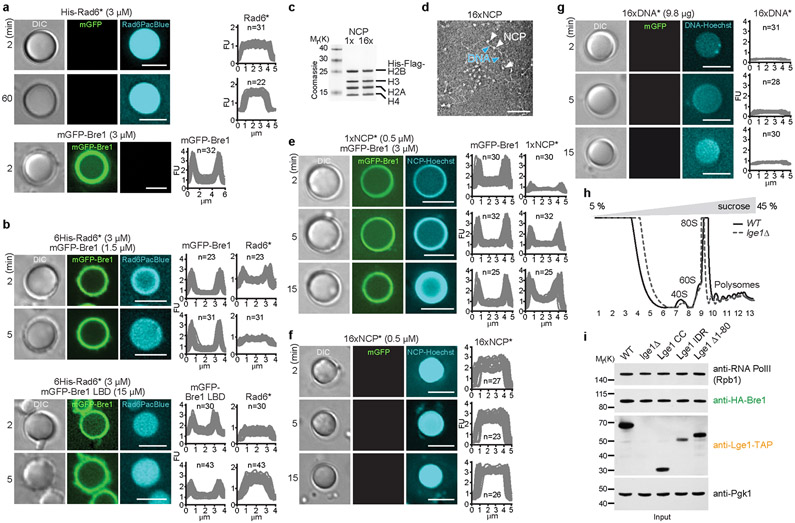
Extended Data Figure 6. Lge1-Bre1 condensates recruit the E2 Rad6 and chromatin.
a. Recruitment of Pacific Blue-labelled 6His-Rad6 (His-Rad6*, 3 μM) or mGFP-Bre1 (3 μM) to Lge1 condensates. Experimental conditions as in Fig. 2a. Fluorescent intensities were quantified across single condensates. FU, arbitrary fluorescent units, n = number of condensates. Scale bar, 5 μm. b. Recruitment of Rad6 to condensates was examined by adding Pacific Blue-labelled 6His-Rad6 (His-Rad6*) to 6His-Lge1 condensates in the presence and absence of an mGFP-Bre1 (1.5 μM) or mGFP-Bre1 LBD (15 μM) shell. The Bre1 LBD construct has a weaker affinity to Lge1, hence requiring a higher concentration. Microscopy was performed immediately after adding His-Rad6* (3 μM) and followed over time. Fluorescent intensities were quantified across single condensates. FU, arbitrary fluorescent units, n = number of condensates. Scale bar, 5 μm. c. Reconstituted mononucleosomes (1x NCP) and oligonucleosomes (16x NCP) were analyzed by SDS-PAGE and Coomassie staining to assess purity and stoichiometry, d. Negative Stain Electron Microscopy was performed to assess the structure of oligonucleosomes. White arrowheads label individual nucleosomes, blue arrowheads the linker DNA. Scale bar, 100 nm. e. Recruitment of mononucleosomes to Lge1 condensates with an mGFP-Bre1 shell. Hoechst-labelled, reconstituted mononucleosomes (1xNCP*; 0.5 μM) were added to 6His-Lge1 condensates with an mGFP-Bre1 shell (3 μM) and imaged over time (min). FU, arbitrary fluorescent units, n = number of condensates. Scale bar, 5 μm. f. Recruitment of oligonucleosomes to Lge1 condensates. Hoechst-labelled, reconstituted 16x nucleosomes (16xNCP*; 0.5 μM) were added to 6His-Lge1 condensates and imaged overtime (min). FU, arbitrary fluorescent units, n = number of condensates. Scale bar, 5 μm. g. Diffusion and retention of 601 Widom DNA into Lge1 condensates. Same setup as in f but with Hoechst-labeled 16x 601 Widom DNA (16xDNA*) added to 6His-Lge1 condensates, n = number of condensates. Scale bar, 5 μm. h. OD 254 nm traces of the sucrose gradient sedimentation assays (5 % - 45 %) show a reproducible fractionation pattern for cell extracts prepared from the indicated strains. The different ribosomal species and fractions are indicated and correspond to the fractions in Fig. 3a. i. Protein levels in cell extracts used for sucrose gradient assays. Anti-Pgk1 serves as a loading control. See SF2 for uncropped gels and western blots.