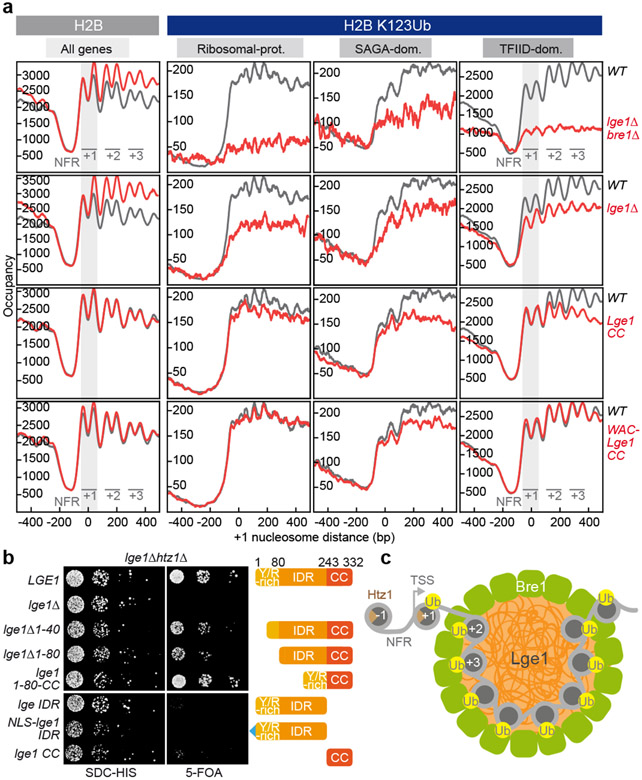
Figure 4. Lge1 IDR enhances H2BK123ub in gene bodies and is required for viability in htz1Δ.
a. H2B (left) or H2BK123ub (right) ChIP-exo tag 5’ ends were plotted relative to the + 1 nucleosome of all genes for WT (gray trace) and the indicated lge1 mutants (red traces). The first three genic nucleosomes are labeled +1, +2 and +3. Two H2B peaks are observed per nucleosome position. H2BK123ub patterns are shown separately for each gene class28. b. Genetic interaction analysis. Double deletion strains harboring a wild-type LGE1 cover plasmid (URA marker) were co-transformed with the indicated plasmids (HIS marker). Growth (10-fold serial dilutions) was followed on SDC-HIS (control) and on SDC+5-fluoroorotic acid (5-FOA), which shuffles out the URA cover plasmid. c. LLPS-based ubiquitination model. The Htz1-containing +1 NCP is depicted outside of the ‘reaction chamber’ as it is ubiquitinated independently of Lge1 LLPS. For simplicity, Rad6 and the E1 are omitted and only a single layer of Bre1 molecules in the shell is shown. TSS, transcription start site; NFR, nucleosome free region.