Malaria parasites grow within circulating red blood cells and uptake nutrients through a pore on their host membrane. Here, we used gene editing to tag CLAG3 and RhopH2, two proteins linked to the nutrient pore, with fluorescent markers and tracked these proteins in living infected cells. After their synthesis in mature parasites, imaging showed that both proteins are packaged into membrane-bound rhoptries. When parasites ruptured their host cells and invaded new red blood cells, these proteins were detected within a vacuole around the parasite before they migrated and inserted in the surface membrane of the host cell. Using simultaneous labeling of CLAG3 and RhopH2, we determined that these proteins interact tightly during migration and after surface membrane insertion. Red blood cells infected with two parasites had twice the protein at their surface and a parallel increase in the number of nutrient pores. Our work suggests that these proteins directly facilitate parasite nutrient uptake from human plasma.
KEYWORDS: FRET, Plasmodium falciparum, fluorescence, malaria, nutrient transport, patch-clamp, protein trafficking
ABSTRACT
Malaria parasites increase their host erythrocyte’s permeability to various nutrients, fueling intracellular pathogen development and replication. The plasmodial surface anion channel (PSAC) mediates this uptake and is linked to the parasite-encoded RhopH complex, consisting of CLAG3, RhopH2, and RhopH3. While interactions between these subunits are well established, it is not clear whether they remain associated from their synthesis in developing merozoites through erythrocyte invasion and trafficking to the host membrane. Here, we explored protein-protein interactions between RhopH subunits using live-cell imaging and Förster resonance energy transfer (FRET) experiments. Using the green fluorescent protein (GFP) derivatives mCerulean and mVenus, we generated single- and double-tagged parasite lines for fluorescence measurements. While CLAG3-mCerulean served as an efficient FRET donor for RhopH2-mVenus within rhoptry organelles, mCerulean targeted to this organelle via a short signal sequence produced negligible FRET. Upon merozoite egress and reinvasion, these tagged RhopH subunits were deposited into the new host cell’s parasitophorous vacuole; these proteins were then exported and trafficked to the erythrocyte membrane, where CLAG3 and RhopH2 remained fully associated. Fluorescence intensity measurements identified stoichiometric increases in exported RhopH protein when erythrocytes are infected with two parasites; whole-cell patch-clamp revealed a concomitant increase in PSAC functional copy number and a dose effect for RhopH contribution to ion and nutrient permeability. These studies establish live-cell FRET imaging in human malaria parasites, reveal that RhopH subunits traffic to their host membrane destination without dissociation, and suggest quantitative contribution to PSAC formation.
INTRODUCTION
Malaria remains a leading infectious cause of morbidity and mortality worldwide; drug resistance in both the Plasmodium pathogen and the mosquito vector threatens to exacerbate this disease burden. The malaria parasite’s success results in part from growth and replication within vertebrate erythrocytes, permitting both evasion of host immunity and access to hemoglobin as a source of amino acids. To enable intracellular development, P. falciparum dramatically remodels its host human erythrocyte by exporting many proteins that alter infected cell cytoadherence, deformability, and permeability to ions and nutrients (1–3).
Increased host cell permeability results from a broad-selectivity ion channel known as the plasmodial surface anion channel (PSAC), which has been linked to the parasite-encoded RhopH protein complex (4, 5). This complex consists of three subunits, termed CLAG, RhopH2, and RhopH3, all of which are conserved throughout Plasmodium spp. infecting primates, rodents, birds, and reptiles (6). While RhopH2 and RhopH3 are expressed from single-copy genes in each species (7, 8), CLAGs are encoded by a multigene family with variable expansion in Plasmodium spp. (9, 10). Two paralogs, CLAG3.1 and CLAG3.2 encoded by nearly identical genes on the P. falciparum chromosome 3, undergo epigenetic switching and have been most confidently linked to PSAC activity through selection of channel mutants, genetic mapping, and DNA transfection studies (11–16).
Rather than synthesis and export to the host membrane immediately before nutrient uptake is initiated, the RhopH proteins are made in the preceding host erythrocyte and packaged into rhoptries, specialized organelles at the apical end of invasive merozoites (17–19). These proteins are then transferred to the next erythrocyte upon invasion and undergo complicated and poorly understood trafficking to the host membrane for channel formation by an unknown mechanism (5, 20). Moreover, because these proteins lack homology to known ion channel proteins, the specific roles served by each subunit remain unclear. While coimmunoprecipitation studies have established interactions between RhopH member proteins, their complicated trafficking through multiple compartments has created uncertainty about whether they remain associated through all parasite developmental stages and subcellular sites. Because one or more subunits may function as chaperones, it is also unclear whether these proteins continue to interact at their final destination, the host erythrocyte membrane.
Here, we explore protein-protein interactions between RhopH subunits using epitope-tagging with GFP derivatives and live-cell imaging (21, 22). We successfully implemented Förster resonance energy transfer (FRET) imaging with fluorescent protein tags on CLAG3 and RhopH2. FRET-positive and -negative control lines confidently implicate sustained interactions between these proteins. These studies provide important insights into an essential parasite activity and should guide structure-function studies of this unusual parasite channel.
RESULTS
GFP derivative tagging for live-cell tracking of RhopH2 and CLAG3.
To establish live-cell tracking of the RhopH complex, we sought to introduce bright GFP derivatives as C-terminal epitope tags on individual subunits of this PSAC-associated complex. We selected two green fluorescent protein (GFP) variants, mCerulean and mVenus (23), with established track records in imaging, including FRET. We then used CRISPR/Cas9 editing to add these tags to RhopH2 and CLAG3 in the P. falciparum KC5 clone, respectively (Fig. 1; see also Fig. S1 in the supplemental material); RhopH3, the third parasite protein linked to PSAC activity, undergoes stage-specific proteolytic processing, rendering C-terminal tagging less effective for trafficking studies (5). Use of the KC5 clone circumvents epigenetic switching between two clag3 genes because this clone carries a single hybrid clag3h gene (24, 25).
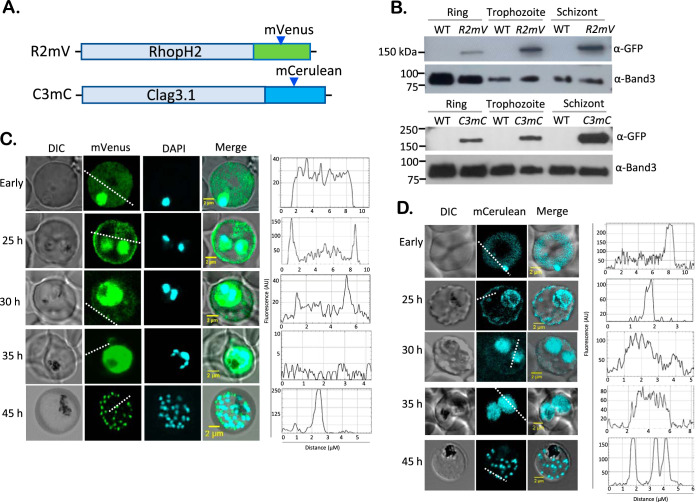
FIG 1.
Stage-dependent trafficking of RhopH2 and CLAG3 in live parasites (A) Ribbon schematics showing C-terminal addition of mVenus and mCerulean tags to rhoph2 and clag3h using CRISPR-Cas9 editing. (B) Immunoblots of indicated parasite clones probed with anti-GFP antibodies that recognize both tags. Lysates harvested at the ring, trophozoite, and schizont stages are shown. Loading control, anti-Band3. Band intensities are not comparable between ring and mature stages. (C) Confocal fluorescence live-cell images of R2mV at the indicated time points after invasion. “Early” indicates infected cells between 0 and 4 h of invasion, as achieved through imaging of live Percoll-enriched schizonts with added erythrocytes. At each stage, line scan analyses are shown in the right panel, based on superimposed dashed white lines. (D) Fluorescence live-cell images of C3mC at indicated time points and corresponding line scan analyses. Images shown for R2mV and C3mC are representative of more than 100 cells visualized over at least 15 experiments.
Production of R2mV and C3mC parasites. (A) CRISPR/Cas9 strategy for tagging of the endogenous rhoph2 gene with mVenus. Site-specific cleavage of rhoph2 by the Cas9-sgRNA complex is repaired through homology-directed repair with homology arms that flank an in-frame mVenus open reading frame. (B) Ethidium stained gels showing PCR integration checks for R2mV and its wild-type KC5 parent. The R2mV clone has fully replaced the wild-type locus with the desired mVenus tag. (C) Full-length R2mV immunoblot, showing the entire blot from Fig. 1B. (D) Strategy for clag3 tagging with mCerulean. (D) Ethidium-stained gels showing PCR integration checks for C3mC and its KC5 parent. Primer positions are as indicated in panel A; primer sequences are provided in Table S1 in the supplemental material. (F) Full-length C3mC immunoblot from Fig. 1B. Download FIG S1, PDF file, 0.3 MB (347.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DNA transfection and limiting dilution cloning yielded recombinant clones, R2mV and C3mC, that fully replaced the wild-type loci (see Fig. S1B and D). DNA sequencing confirmed in-frame addition of the GFP derivatives in both parasites. Immunoblotting with anti-GFP antibodies recognized single bands of expected size, indicating stable expression of epitope-tagged proteins (Fig. 1B). Microscopic examination of these clones showed normal progression through the intracellular parasite cycle (Fig. 1C and D, differential interference contrast images), consistent with little or no detrimental effect of the added tags.
We then used live-cell fluorescence imaging to determine the stage-specific locations of each protein (Fig. 1C and D, mVenus and mCerulean columns). Interestingly, at early time points after erythrocyte invasion, both RhopH2 and CLAG3 exhibited a diffuse pattern of fluorescence within the host erythrocyte compartment, as well as an intense signal associated with the intracellular parasite, presumably reflecting protein within the parasitophorous vacuole prior to export via the PTEX translocon (5, 26) (“early” time points reflecting 2 to 4 h after erythrocyte invasion, Fig. 1C and D). As these parasites matured, a rim fluorescence pattern became increasingly apparent, suggesting completed trafficking and insertion in the host erythrocyte membrane (25-h time point). Intensity line scans confirmed an increasingly peripheral localization as the parasite developed into hemozoin-containing trophozoites (dashed lines and corresponding intensity plots at right, Fig. 1C and D). Late within the cycle (schizont stage, 45 h), both clones exhibited bright punctate patterns that reflect trafficking of newly synthesized RhopH proteins to daughter merozoite rhoptries for secretion into the next erythrocyte upon egress and reinvasion. These live-cell images provide insights into the trafficking of two RhopH member proteins from the parasitophorous vacuole into the host cytosol and eventually to the host membrane.
Doubly infected cells exhibit parallel increases in RhopH2 export and PSAC activity.
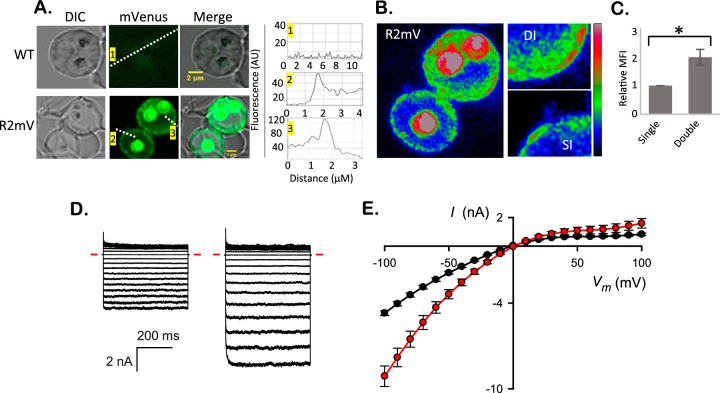
In vitro cultures contain occasional erythrocytes infected with multiple parasites because two or more merozoites can successfully invade and grow within a single host cell. We sought and found R2mV double-infected cells and then used nearby single-infected cells to normalize the total fluorescence intensities from these cells (Fig. 2A and B). Using the adjacent single-infected cells to control for photobleaching and other imaging artifacts, we found significantly higher fluorescence signals in double-infected cells (Fig. 2C; P = 0.04, paired t test, n = 3 cell pairs). Because RhopH2 is produced in the preceding erythrocyte cycle and delivered to the host cell upon reinvasion (5, 20), the ∼2-fold-higher fluorescence in double-infected cells suggests that RhopH2 transfer is not compromised when more than one merozoite invades an erythrocyte.
FIG 2.
Double-infected cells exhibit parallel increases in fluorescence and permeability. (A) Live-cell images and line scan analysis of WT and R2mV parasites at 25 h after invasion. Note the cell infected with multiple WT parasites produces negligible fluorescence and that the signal from two R2mV parasites is greater than from the adjacent single-infected cell. Numbered line scans showing intensity differences are show in the right panels. (B) Rainbow-scaled image of the infected cells from panel A, with color indicating mVenus fluorescence intensity. Right, zoom in images of the host membrane from these single- and double-infected cells (SI and DI). Color scale bar at right, 14 to 210 AU linear gradient. (C) Mean fluorescence intensities (MFI) ± the SEM from matched pairs of adjacent cells infected with one or two R2mV parasites, determined using whole infected cells as the defined regions of interest. Double-infected cells have increased total fluorescence intensities. *, P = 0.04 (n = 3 cell pairs). (D) Ensemble presentation of whole-cell currents in response to voltage pulses between −100 mV and +100 mV in 10-mV increments. Left and right traces are from single- and double-infected cells, respectively. The inward-rectifying currents are characteristic of PSAC activity (27). Red dashes, zero current level. (E) Mean currents (I) ± the SEM at a range of applied membrane potentials (Vm) determined from 23 single- and 10 double-infected erythrocytes (black and red symbols, respectively).
To explore the functional consequences of increased RhopH protein in the host compartment of double-infected cells, we performed whole-cell patch-clamp and compared PSAC-mediated currents. Although whole-cell currents vary from cell-to-cell, we found consistently larger currents on cells carrying two parasites (Fig. 2D and E). Chord conductances, measured between −50 and −100 mV, were significantly greater (115 nS ± 8 versus 56 ± 2 for single-infected cells, n = 10 and 23 cells, respectively, P < 10−4). Notably, these conductances matched the 2-fold increase in RhopH2 transfer, implicating a linear dose relationship between RhopH2 protein delivery to host cells and channel-mediated permeability. Both single- and double-infected cells exhibited inward rectifying currents characteristic of PSAC; when combined with noise analysis and pharmacological studies (12, 27), this observation has established that PSAC is the predominant conductive pathway on the infected host cell membrane. Thus, increased RhopH2 delivery yields higher ion permeabilities by increasing the functional PSAC copy number on the host membrane.
Tandem labeling establishes that RhopH subunits colocalize throughout the parasite cycle.
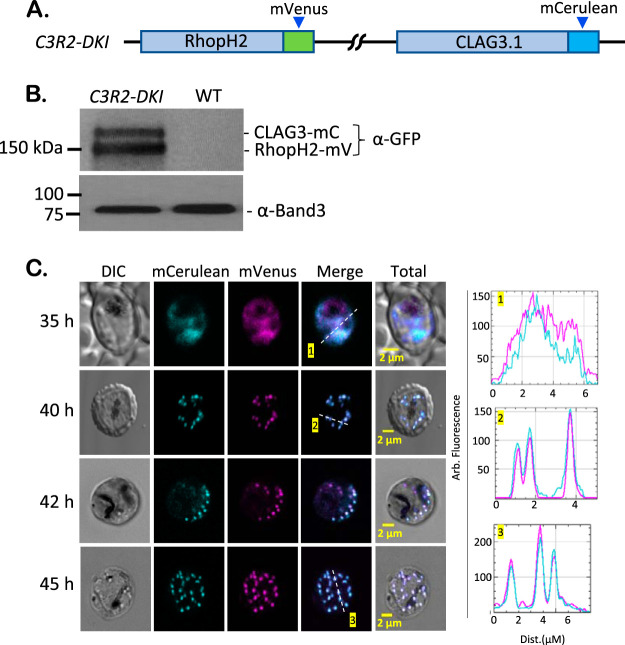
To examine associations between these conserved proteins, we then performed sequential CRISPR transfections to generate a parasite with simultaneous fluorescent tagging of RhopH2 and CLAG3. The R2mV clone was transfected with pL6-Clag3h-mC; selection with WR99210 and subsequent limiting dilution cloning yielded C3R2-DKI, the desired double-tagged parasite (Fig. 3A; see also Fig. S2A). PCR and DNA sequencing confirmed successful production of this integrant (see Fig. S2B). Immunoblotting with GFP antibodies that recognize both the mVenus and the mCerulean derivatives revealed two distinct bands that migrated at approximately 150 and 175 kDa (Fig. 3B), somewhat smaller than predicted by the sequences of the engineered proteins; prior studies have also observed aberrant migration of these membrane-associated proteins (25). Importantly, the doublet observed in C3R2-DKI, but not in the R2mV and C3mC single-tag clones (Fig. 1B), confirms that both RhopH2 and CLAG3 were successfully tagged. We then used imaging to track the movement of both proteins within individual parasites at various intracellular parasite stages and established that these proteins colocalize at all examined stages within the infected erythrocyte (Fig. 3C).
FIG 3.
Strict association between RhopH2 and CLAG3 revealed by imaging tandem-labeled cells. (A) Ribbon diagram showing tandem labeling of RhopH2 and Clag3h through sequential CRISPR-Cas9 transfection to produce the C3R2-DKI clone. (B) Immunoblot of total cell lysates from indicated parasites, probed with anti-GFP antibody. Two bands are detected in C3R2-DKI, consistent with tagging of both CLAG3 and RhopH2. Loading control, Band3. (C) Live-cell images of C3R2-DKI-infected cells at the indicated time points after invasion. RhopH2 and CLAG3 colocalize within trophozoites (top row) and within rhoptries of mature schizonts (other rows). Right panels, corresponding line scans. The results are representative of >80 cells imaged from at least eight experiments.
Sequential transfections to produce double-tagged parasites. (A) Flowchart showing strategy for production of C3R2-DKI. (B) Ethidium-stained gels showing PCR integration checks for C3R2-DKI and its KC5 parent. (C) Full-length immunoblot from Fig. 3B. (D) Strategy for production of R2mV-LmC. (E) PCR confirming preserved integrated of the mVenus tag on rhoph2 and retention of the episome for expression of mCerulean with a RhopH2 leader sequence in R2mV-LmC parasites. Download FIG S2, PDF file, 0.3 MB (298.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
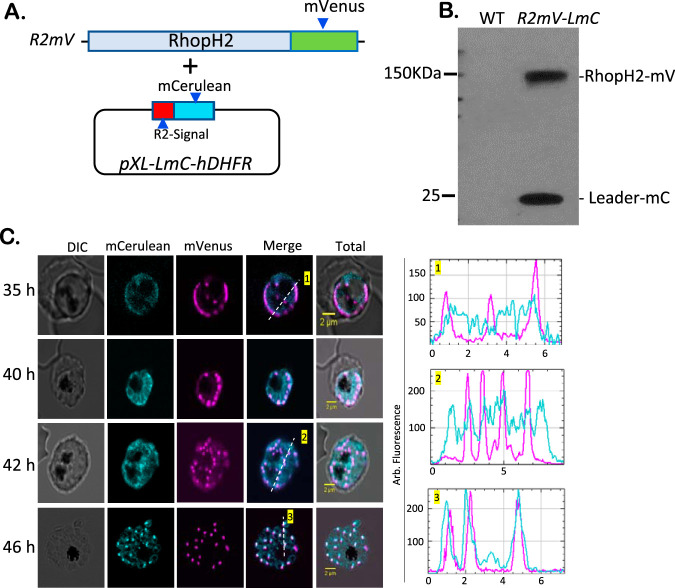
A control double-labeled parasite with distinct trafficking kinetics.
We next produced R2mV-LmC, a FRET negative-control parasite, that targets the two GFP derivatives to the parasite rhoptry without specific protein-protein interactions. We transfected R2mV with pXL-LmC-hDHFR, an episome that expresses mCerulean behind the 24-amino-acid (aa) N-terminal sequence of RhopH2 (Fig. 4A). This leader sequence has previously been used to target GFP to the parasite rhoptries (19). Immunoblotting with anti-GFP revealed an additional lower-molecular-weight band that corresponds to the leader sequence mCerulean chimera, indicating that both GFP derivatives were successfully expressed (Fig. 4B). Stage-specific imaging of R2mV-LmC confirmed coexpression and revealed a surprising difference in trafficking kinetics. At the late trophozoite stage, the two GFP derivatives were comparably expressed and were diffusely distributed throughout the parasite (35 h, Fig. 4C). However, at 40 to 42 h after invasion, these reporters exhibited distinct subcellular distributions. The full-length RhopH2 tagged with mVenus exhibited a punctate pattern, suggesting completed trafficking to rhoptries soon after these organelles have developed. In contrast, mCerulean, expressed behind only a short leader sequence, remained diffusely distributed (40 and 42 h, Fig. 4C); this smaller protein eventually trafficked to the rhoptry and colocalized with mVenus (46 h). Thus, although we confirmed that the leader sequence is sufficient to traffic GFP derivatives to the rhoptry (19), coexpression and imaging of tightly synchronized cultures revealed measurably slower trafficking kinetics than for the tagged native protein. We propose that trafficking of full-length RhopH2 is facilitated by its association with CLAG3 and RhopH3. Consistent with this proposal, these three proteins undergo cotranslational assembly prior to transit, with knockdown of RhopH3 compromising transit of other subunits to the rhoptry (5).
FIG 4.
Distinct trafficking kinetics in R2mV-LmC, a control tandem-labeled parasite. (A) Schematic showing transfection of R2mV with the pXL-LmC-hDHFR plasmid for episomal expression of mCerulean behind a 24-aa N-terminal RhopH2 leader sequence. (B) Anti-GFP immunoblot showing two expected bands in the R2mV-LmC parasite. (C) Fluorescence imaging of R2mV-LmC at the indicated time points after invasion. Note that the full-length mVenus-tagged RhopH2 protein traffics to developing rhoptries rapidly to yield punctate labeling, but mCerulean expressed behind a short leader sequence exhibits delayed trafficking. Right panels, color-coded line scans from corresponding images. The results are representative of >50 cells imaged in at least four experiments.
Live-cell FRET imaging establishes sustained association between CLAG3 and RhopH2.
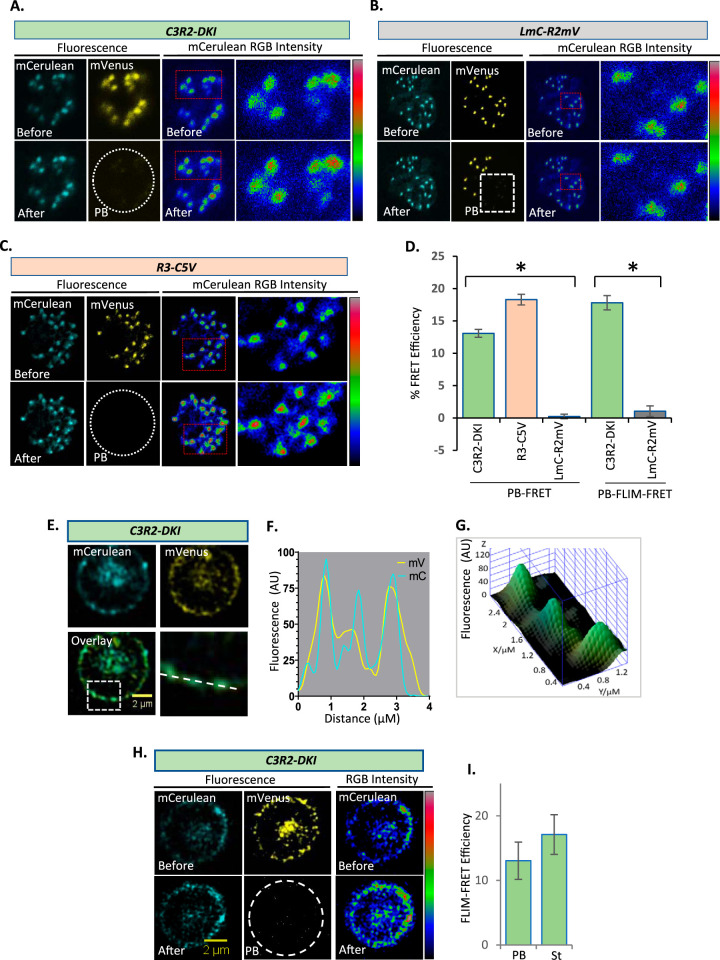
Although colocalization studies have suggested association between the RhopH complex members at each parasite stage (5), fluorescence microscopy as used in these studies is limited by the low spatial resolution afforded by the Abbe diffraction limit. For small organelles such as the rhoptry, this resolution limit may incorrectly implicate colocalization and protein-protein interactions, as exemplified by the two noninteracting reporter proteins in R2mV-LmC (Fig. 4C, 46-h time point). Live-cell FRET studies can quantify protein-protein interaction and can directly address this concern (28, 29). We selected acceptor photobleaching and fluorescence lifetime imaging (FLIM) to measure FRET efficiency between mCerulean and mVenus in C3R2-DKI to assess association between CLAG3 and RhopH2. In schizont-stage parasites, where these proteins are synthesized and packaged into rhoptry organelles, photobleaching of the RhopH2-mVenus acceptor produced a visually apparent increase in CLAG3-mCerulean fluorescence (Fig. 5A), as expected from donor dequenching.
FIG 5.
FRET between CLAG3 and RhopH2 at distinct sites within infected cells. (A) Live-cell fluorescence microscopy of a C3R2-DKI schizont-infected cell before and after acceptor photobleaching (PB). Right panels show RGB color scaling of mCerulean fluorescence intensity; increased red labeling of individual rhoptries in the bottom panels indicates increased mCerulean intensity after mVenus photobleaching. Color scale bar at right, 4 to 103 AU linear gradient. (B and C) Identical photobleaching experiments with LmC-R2mV negative-control parasite (RGB scale bar, 6 to 163 AU) and R3-C5V positive-control parasites (RGB scale bar, 0 to 298 AU). Images representative of 31 to 151 cells for each line. (D) Mean FRET efficiencies ± the SEM for indicated parasites using photobleaching FRET (PB-FRET; calculated from the donor mean fluorescence intensity [MFI] pre- and post-acceptor photobleaching) or PB-FLIM-FRET efficiencies (calculated from donor lifetimes pre- and post-acceptor photobleaching). *, P < 10−4. (E) Host membrane colocalization of CLAG3-mCerulean and RhopH2-mVenus in a trophozoite-stage C3R2-DKI-infected cell. Colocalization at the host membrane is highlighted (boxed region and dashed line). (F) Corresponding line scan with yellow and cyan lines representing mVenus and mCerulean intensities. (G) Three-dimensional surface plot from the along the dashed line in panel F. (H) Live-cell images of trophozoite-stage C3R2-DKI parasite before and after acceptor photobleaching to estimate FLIM-FRET. Right panels, RGB color scaling of mCerulean intensity (scale bar, 77 to 125 AU). (I) Mean FLIM-FRET efficiencies ± the SEM at the host membrane using C3R2-DKI parasites, calculated using both photobleaching and standard methods (PB and St, respectively; n = 2 cells each).
Because FRET results may also be affected for proteins with restricted mobilities due to confined proteins environments or membrane association (30), we used the R2mV-LmC as a negative control for our FRET measurements. Figure 5B shows that acceptor photobleaching in this control did not clearly produce donor dequenching. We also engineered a positive-control line R3-C5V, where both the mCerulean donor and the mVenus acceptor are appended to RhopH3 and separated by only 5 aa (Fig. S3). This arrangement has been shown to produce robust FRET in mammalian cells (31). PCR, DNA sequencing, and immunoblotting confirmed production of this parasite and revealed that this large-tandem tag permits imaging studies in mature schizonts, but not other stages due to stage-specific proteolytic processing of the C-terminal tag (Fig. S3B and C). Acceptor photobleaching in this line produced donor dequenching as expected (Fig. 5C).
R3-C5V FRET positive-control parasite. (A) CRISPR/Cas9 strategy. (B) PCR integration checks for the R3-C5V clone and its wild-type KC5 parent. Primer sequences are in Table S1 in the supplemental material. (C) Immunoblots probed with anti-GFP showing absence of signal in untransfected wild-type cells (WT) and the tandem mCerulean-mVenus tagged RhopH3 protein (band at 150 kDa) in R3-C5V parasites. The blot on the right shows a stage-specific cleavage product at 45 kDa that results from proteolytic processing of RhopH3 in parasite rhoptries (5). Download FIG S3, PDF file, 0.3 MB (366.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
We quantified FRET efficiency in these experiments with the energy transfer equation E = (FD – FDA)/FD =1 – (FDA/FD), where FDA and FD represent the donor signal intensity before and after acceptor photobleaching, respectively. This revealed a maximal FRET efficiency under our experimental conditions of 18.29% ± 4.25% based on the strictly associated donor-acceptor pair in R3-C5V (Fig. 5D, PB-FRET). The negative control, R2mV-LmC, yielded negligible FRET, establishing that these measurements can identify noninteracting protein pairs in parasite rhoptries. For the C3R2-DKI line, schizont-stage parasites yielded significant photobleaching FRET (P < 10−4, n = 31 to 151 cells each, one-way analysis of variance [ANOVA] with a post hoc Tukey’s multiple-comparison test). These findings strongly support molecular association between CLAG3 and RhopH2 within parasite rhoptries without the caveat of measurements in confined spaces. Because these efficiencies approach those of the positive-control R3-C5V, we propose that CLAG3-RhopH2 association is quantitative with nearly all complexes remaining intact during traffic from their site of synthesis to their eventual destination at the host erythrocyte membrane.
We also performed FRET using fluorescence lifetime imaging microscopy (FLIM-FRET) to examine association between CLAG3 and RhopH2. Typically, FLIM-FRET is measured by examining the lifetime of the donor alone and comparing that to the lifetime when the donor is associated with the acceptor (standard FLIM-FRET). Alternatively, FLIM-FRET can be measured by comparing the lifetime of the donor in the presence of the acceptor before and after photobleaching (PB-FLIM-FRET). We employed both methods to measure donor lifetime (32). For schizont-stage parasites, we examined mCerulean donor lifetime in C3R2-DKI before and after photobleaching of the mVenus acceptor. We calculated FLIM-FRET efficiency using E = 1 – τ/τ0, where τ and τ0 are the lifetimes of the donor fluorescence before and after acceptor photobleaching, and found an efficiency comparable to that observed above with the standard photobleaching FRET method (Fig. 5D, PB-FLIM-FRET and PB-FRET, respectively). We also examined PB-FLIM-FRET in the R2mV-LmC negative-control parasite and found negligible FRET, confirming that the detected changes in donor lifetime upon acceptor photobleaching corresponds to a stable interaction between CLAG3 and RhopH2 within rhoptries (P < 10−4, n = 15 to 22 regions of interest each from separate cells, one-way ANOVA with a post hoc Tukey’s multiple-comparison test).
Because FLIM-FRET approaches permit sensitive detection of FRET at lower fluorophore concentrations than needed for fluorescence intensity-based FRET, it was also possible to use lifetime measurements for the lower abundance protein at the host membrane of trophozoite-infected cells (Fig. 5E and G to I). In addition to the acceptor photobleaching approach used above, we used standard FLIM-FRET. Here, mCerulean lifetime was measured in C3R2-DKI parasites without photobleaching and compared to its lifetime in control C3mC parasites, which lack the mVenus acceptor.
Although reduced fluorescence signals of mCerulean at the host membrane limited these analyses to three trophozoite-stage C3R2-DKI-infected cells, we obtained comparable FLIM-FRET efficiencies with both photobleaching and standard approaches (Fig. 5I, PB and St, respectively). We suspect that a combination of factors—low efficiency of parasite protein delivery to the host membrane, a greater abundance of RhopH2 than CLAG3 at the host membrane because of competition with other CLAG paralogs for trafficking of the protein complex, and the lower brightness of the mCerulean donor than of the mVenus acceptor—limited the number of cells with suitable mCerulean signals at the host membrane. Most importantly, the FRET efficiencies we estimated using two FLIM-FRET methods were in good agreement with efficiency estimates from the larger number of schizont-stage cells we analyzed (Fig. 5D). These findings implicate quantitatively sustained interaction between CLAG3 and RhopH2 at the host erythrocyte membrane.
DISCUSSION
Malaria parasites export many effector proteins into the erythrocytes they infect (33, 34). A prime example of an essential exported activity is the plasmodial surface anion channel (PSAC), a broad selectivity channel that permits nutrient acquisition by the intracellular pathogen (3). Although members of the RhopH complex have been linked to PSAC with several independent approaches (4, 13, 16), important questions remain about how these proteins directly or indirectly induce channels at the host membrane (35). Here, we have used tagging of two RhopH member proteins, RhopH2 and CLAG3, to examine stage-specific trafficking and protein-protein interactions in live parasite-infected cells and make several useful insights.
The RhopH complex of three proteins was initially identified through immunoprecipitation studies that date to the 1980s (36–38). More recently, molecular and cell biological studies have implicated trafficking of these proteins from the rhoptry into the parasitophorous vacuole before transfer into host cytosol and insertion into the host membrane for channel formation (5, 20). Because each member protein exists in forms that are either peripheral or integral to membranes (5), there appear to be distinct pools of RhopH proteins that may vary from one subcellular location to another. Then, coimmunoprecipitation from cellular lysates may reflect association at only specific locations, casting doubt on whether these proteins continue to interact at all cellular sites and developmental stages. Importantly, if RhopH2 and/or RhopH3 function only as chaperones that deliver CLAG3 to the host membrane for channel formation, these proteins may not remain associated once they reach the host cell membrane. In this scenario, colocalization at the host membrane, as observed in confocal immunofluorescence microscopy (20), could be misleading. We address this concern with FRET studies, providing experimental evidence for sustained association between RhopH2 and CLAG3 at their final membrane destination.
To our knowledge, this is the first study to use live-cell FRET in malaria parasites to examine protein-protein interactions. Successful demonstration required production of a double-tagged parasite clone and was enabled through sequential CRISPR transfection; we also used transfection to generate double-tagged positive and negative controls to address theoretical concerns and quantify FRET efficiency. While sequential tagging of two proteins was formally possible with previous single-crossover transfections (39, 40), frequent loop-out of those plasmids produces mixed populations of parasites, preventing rigorous examination and stage-specific tracking. Prior to implementation of CRISPR/Cas9 editing in P. falciparum (41), FRET measurements in malaria were limited to biosensor molecules to detect interactions with protein targets (42–44) and fixed-cell assays with tagged antibodies (45, 46).
FRET studies represent a powerful method for examining molecular-level interactions between proteins. Nevertheless, study of membrane-associated proteins or proteins that localize to small organelles such as rhoptries raises concerns about false-positive FRET signals resulting from transient and random protein collisions in small spaces (47, 48). Another limitation is that it is often critical to maintain protein expression at physiologically relevant levels to ensure that observed interactions are biologically meaningful (49). R2mV-LmC, a separate double-tagged parasite that targets mCerulean to rhoptries without CLAG3 sequences that mediate RhopH2 interaction, was used to examine this limitation. Although the minimal leader sequence with mCerulean reporter and the full-length RhopH2 with a C-terminal mVenus tag colocalize within rhoptries, this R2mV-LmC parasite exhibited negligible FRET (Fig. 5), providing a definitive negative control and suggesting that FRET measurements can be reliably made with these proteins despite their membrane association and confinement to rhoptries. Compartmentalization and/or reduced mobility of proteins within this organelle, which could additionally complicate interpretation of FRET studies, should be studied as the required technologies become available.
The R2mV-LmC clone also provides new insights into how RhopH proteins traffic to rhoptries after their synthesis. Prior studies have implicated cotranslational assembly of the RhopH complex (5) and conventional trafficking to rhoptries via endoplasmic reticulum and Golgi structures (50). Here, we extend this finding by showing that the RhopH2 leader sequence can adequately target mCerulean to the rhoptry but that this protein exhibits delayed trafficking compared to full-length RhopH2. Simultaneous imaging of the two fluorescent reporters in individual cells circumvented problems arising from differing developmental stages for parasites within a culture and enabled direct comparison. With this insight, we propose that the association between RhopH proteins facilitates efficient trafficking to the rhoptry.
Upon merozoite egress and invasion of new erythrocytes, RhopH proteins are deposited into the parasitophorous vacuole (early time points, Fig. 1C and D). Although we did not perform FRET studies to examine continued interaction in the C3R2-DKI line at this stage, it seems likely that the RhopH subunits are associated upon initial deposit in this vacuole. The complex must then cross the vacuolar membrane to enter the host cell; it is unclear whether this export occurs via the PTEX translocon (5, 26). If a translocon is involved, the complex presumably undergoes transient dissociation to allow protein export. After export, these proteins reassociate and traffic to the host membrane, possibly facilitated by exported chaperones and membranous organelles known as the Maurer’s clefts (51, 52). Although FRET studies can, in principle, be used to examine transient dissociation and reassociation of the RhopH complex as it crosses the vacuolar membrane, such imaging studies will be complicated by the large pool of intact RhopH complexes within the vacuole and rate-limited export at this membrane.
When these proteins finally reach their host membrane destination, RhopH2 and CLAG3 are stably associated, as now established by our studies. Along with inhibitor and protease susceptibility studies (14), our finding that PSAC-mediated whole-cell currents increase linearly with RhopH2 abundance in double-infected erythrocytes suggests that these proteins contribute directly to formation of the nutrient uptake channel. Functional studies have also revealed quantitative reduction in PSAC currents with RhopH2 and RhopH3 knockdown (5); interestingly, CLAG3 knockdown and knockout produce unexpectedly modest effects on channel phenotypes (53). A clearer understanding of how these proteins contribute to channel-mediated nutrient uptake will facilitate antimalarial drug development targeting this essential parasite activity (12). It would also enable structural insights into how this channel permits uptake of diverse nutritive solutes without compromising the osmotic stability of the host erythrocyte (54). Reporter parasites such as the C3R2-DKI line should enable these insights and may also prove useful for identification of drug leads that interfere with assembly or trafficking of the RhopH complex.
MATERIALS AND METHODS
Parasite cultures.
The asexual P. falciparum KC5 parental line and transfectant clones generated from it were confirmed by DNA sequencing and were cultivated in O+ human erythrocytes (Interstate Blood Bank, Inc.) using standard methods. Cultures were maintained at 5% hematocrit under 5% O2, 5% CO2, and 90% N2 at 37°C.
When required for imaging specific parasite developmental stages, cultures were tightly synchronized with a two-step protocol. Synchronous schizont-stage infected cells were enriched with the Percoll-sorbitol method, returned to culture for 4 to 6 h to allow egress, and subjected to treatment with 5% sorbitol to remove schizonts that had not matured to egress. This procedure yields a homogeneous population of early ring-stage parasites that can be followed through the intracellular cycle. Immunoblots were performed using cultures subjected to two rounds of treatment with 5% sorbitol; examination of Giemsa-stained smears was then used to confirm that parasites were predominantly at the indicated stages.
Plasmid design and construction.
The mVenus C1, mCerulean C1, and C5V constructs were gifts from S. Vogel (Addgene plasmid numbers 26394, 27794, and 27796) (31).
A two-plasmid CRISPR/Cas9 strategy was used to produce parasites expressing CLAG3 and/or RhopH2 proteins carrying C-terminal fluorescent tags (5). CRISPR guide RNAs were selected based on specific cleavage of target genes near the C terminus with consideration of high on-target efficiency scores (55, 56). The pL6-rhopH2-mVenus plasmid was constructed by restriction digestion of pL6-rhopH2-PA-glmS with Kas-I and NheI and subsequent ligation of mVenus amplified with appropriate overhangs (New England Biolabs, primers MA-mV/F1 and MA-mV/R2; Table S1) (5). pL6-Clag3-mCerulean was constructed in two steps. First, In-Fusion cloning was used to assemble a cassette consisting of 5′ and 3′ homology arms and an intervening recodonized terminal clag3 fragment with CRISPR shield mutations; the resulting DNA was amplified using primers Inf-C3-5HA-F1 and Inf-C3-3HA-R1 and inserted into the pL6 backbone. Subsequently, mCerulean was amplified using the primers Inf-C3-mC-F1 and Inf-C3mC-R1 and inserted in-frame at unique KasI and NheI sites. The pL6-rhopH3-C5V plasmid was generated by subcloning a C5V amplicon (primers Inf-R3C5V-OF1 and Inf-R3C5V-OR1) into pL6-rhopH3-PA-glmS (5) at unique NheI and NarI sites. CRISPR guide RNAs were cloned through In-Fusion cloning of annealed complementary oligonucleotides (TaKaRa; sgRNAs are listed in Table S1).
Primers and sgRNAs used in this study. Download Table S1, XLSX file, 0.01 MB (12.3KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The FRET negative-control parasite, R2mV-LmC, was produced by transfection of R2mV parasites with the pXL-LmC-hDHFR plasmid for episomal expression of mCerulean with a RhopH2 leader sequence. A double-stranded DNA construct encoding the 24-aa N terminus of RhopH2, the 4-aa linker, and mCerulean was synthesized and cloned into a pXL plasmid behind a parasite msp2 promoter for stage-specific expression similar to that of rhoph genes (25).
DNA sequencing and restriction digestion were used to confirm all constructed plasmids.
P. falciparum DNA transfections.
Electroporation was used to load 175 μl packed uninfected erythrocytes with 50 μg of each required plasmid in 1 ml of cytomix (120 mM KCl, 0.15 mM CaCl2, 2 mM EGTA, 5 mM MgCl2, 10 mM K2HPO4, 10 mM KH2PO4, 25 mM HEPES [pH 7.6]; Gene Pulser Xcell system; Bio-Rad Laboratories). Percoll-sorbitol enriched trophozoite-infected cells were added and cultivated for 24 h before addition of 1.5 μM DSM1 for ydhodh selection (BEI Resources); sequential knock-in to produce C3R2-DKI used 2.5 nM WR99210 for hdhfr selection (Jacobus Pharmaceuticals). Selection for retention of the pXL-LmC-hDHFR episome also used WR99210. After 20 to 25 days, viable parasites were detected by Giemsa-stained microscopy. Integration PCR checks were performed using genomic DNA isolated from parasite. Integrant parasite lines were cloned by limiting dilution for these studies.
Electrophysiology.
Trophozoite-infected erythrocytes (Dd2 and TFLC3 strains) were used for patch-clamp studies; these strains produce comparable increases in permeability and whole-cell currents (25). Whole-cell patch-recordings were obtained using symmetric pipette and bath solutions of 1 M choline chloride, 115 mM NaCl, 10 mM MgCl2, 5 mM CaCl2, 20 mM Na-HEPES (pH 7.4) and quartz pipettes (27). Erythrocytes were identified as infected with either one or two parasites based on visual examination of hemozoin and selected for formation of gigaseals (typically, >100 GΩ). Current responses to applied membrane potentials between −100 and +100 mV were acquired using Clampex 10.0 software and an Axopatch 200B amplifier (Molecular Devices), low pass filtered at 5 kHz (8-pole Bessel filter; Frequency Devices), and digitized at 100 kHz using a Digidata 1550B converter (Molecular Devices).
Live imaging and FRET methods.
Live images of all the recombinant parasites were collected on Laser scanning microscope 880 (LSM880; Zeiss) equipped with a high-quantum-efficiency GaAsP detector that permits low-laser-power excitation and minimal phototoxicity. All images were acquired at 37°C using preincubation to minimize focus drift. A Plan-Apochromat 63 × 1.4 NA oil-immersion lens objective was used for FRET imaging. mVenus and mCerulean were excited at 514 nm and at 405 or 440 nm, respectively. Cross talk between these fluorescent proteins in double transfectants was minimized by adjusting emission bandpass settings and confirmed using control measurements using parasites that express individual reporter proteins. DAPI (4′,6′-diamidino-2-phenylindole) was not included as a nuclear marker in mCerulean imaging studies because of cross talk. Images were processed using Zeiss Zen or FIJI (ImageJ). Line scans were performed using standardized intensity settings, yielding values that, although presented in arbitrary units, are comparable between cells.
FRET imaging used several complementary methods. Photobleaching FRET was performed by photobleaching the acceptor protein to measure the extent of donor dequenching. Time series were set to acquire images pre- and postbleaching of the acceptor mVenus with a 514-nm laser set to 100%. A threshold of >95% acceptor photobleaching was implemented for accurate FRET efficiency calculations.
For standard and photobleaching FLIM-FRET measurements, we used a Becker & Hickl FLIM system coupled with a Zeiss LSM 780 laser scanning confocal microscope equipped with a pulsed TiS laser (Chameleon; Coherent) and ZEN (Carl Zeiss) acquisition software. Briefly, our FLIM workstation is equipped with two Becker & Hickl FLIM GaAsP hybrid detectors, a DCC-100 detector controller module, two SPC-150 time-correlated single photon counting modules, and a separate computer workstation to control the FLIM detectors and module boards. The FLIM hardware is not controlled through the ZEN software. For data acquisition, laser scanning parameters are set and started in ZEN, with photon counting accomplished by using Becker & Hickl FLIM software.
For standard FLIM-FRET measurements, the donor-only C3mC parasite was used to measure the donor fluorophore lifetime. The C3R2-DKI donor-acceptor pair and C3mC donor only parasites were imaged by exciting the mCerulean donor at 405 nm. Lifetimes were estimated using SPCImage software (Becker & Hickl) and used for FRET efficiency calculations.
For acceptor-photobleaching (PB)-FLIM-FRET, mCerulean lifetime was measured before and after mVenus photobleaching of C3R2-DKI parasites. FLIM data were analyzed using SPCImage software. Data were then exported to Microsoft Excel and SigmaPlot 10.0 for statistical analyses.
Immunoblots.
Immunoblotting was performed using cultivated synchronous parasites diluted with hypotonic lysis buffer (10 mM Tris-HCl, 1 mM phenylmethylsulfonyl fluoride [pH 7.5]). While trophozoite- and schizont-stage cultures were enriched using Percoll-sorbitol, this method is not effective for immature ring-stage cultures, which were instead harvested at high parasitemias (>10%). Thus, direct comparisons of protein levels between ring-infected and mature parasites cannot be made. Total cell lysates were reduced in a modified Laemmli sample buffer containing 6% SDS. Proteins were resolved by electrophoresis in a 4 to 15% Mini-Protean TGX gel (Bio-Rad) and transferred to nitrocellulose. After blocking with 3% skim milk in 150 mM NaCl–20 mM Tris-HCl (pH 7.4) with 0.1% Tween 20, anti-GFP rabbit polyclonal primary antibody (Clontech) was used to detect both mVenus and mCerulean epitope tags and was applied at a 1:4,000 to 1:5,000 dilution in blocking buffer overnight at 4°C. Anti-Band3 antibody (Thermo Fisher Scientific) was similarly used at a 1:5,000 dilution to detect the Band3 loading control. After washing, horseradish peroxidase-conjugated secondary antibodies (Sigma-Aldrich) were applied at a 1:2,000 dilution for 2 h at room temperature. After washing, chemiluminescent substrate (Clarity Western ECL substrate; Bio-Rad) was typically applied; immunoblots for C3R2-DKI lysates used Sigma FAST DAB with urea/metal enhancer to improve detection of the two GFP derivatives on directly on the membrane. All blots are representative of n = 3 to 4 independent trials.
Statistical analysis.
Numerical data were tallied and plotted as means ± the standard errors of the mean (SEM). The statistical significance was calculated by using an unpaired Student t test or one-way ANOVA with post hoc testing, as indicated. Significance was accepted at P < 0.05.
ACKNOWLEDGMENTS
We thank D. Jacobus and BEI resources for providing WR99210 and DSM-1 for parasite transfections.
This study was supported by the Intramural Research Program of National Institutes of Health, National Institute of Allergy and Infectious Diseases. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Citation Ahmad M, Manzella-Lapeira J, Saggu G, Ito D, Brzostowski JA, Desai SA. 2020. Live-cell FRET reveals that malaria nutrient channel proteins CLAG3 and RhopH2 remain associated throughout their tortuous trafficking. mBio 11:e01354-20. https://doi.org/10.1128/mBio.01354-20.
REFERENCES
- 1.Boddey JA, Cowman AF. 2013. Plasmodium nesting: remaking the erythrocyte from the inside out. Annu Rev Microbiol 67:243–269. doi: 10.1146/annurev-micro-092412-155730. [DOI] [PubMed] [Google Scholar]
- 2.Cyrklaff M, Sanchez CP, Frischknecht F, Lanzer M. 2012. Host actin remodeling and protection from malaria by hemoglobinopathies. Trends Parasitol 28:479–485. doi: 10.1016/j.pt.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Desai SA. 2014. Why do malaria parasites increase host erythrocyte permeability? Trends Parasitol 30:151–159. doi: 10.1016/j.pt.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguitragool W, Bokhari AA, Pillai AD, Rayavara K, Sharma P, Turpin B, Aravind L, Desai SA. 2011. Malaria parasite clag3 genes determine channel-mediated nutrient uptake by infected red blood cells. Cell 145:665–677. doi: 10.1016/j.cell.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito D, Schureck MA, Desai SA. 2017. An essential dual-function complex mediates erythrocyte invasion and channel-mediated nutrient uptake in malaria parasites. Elife 6:e23485. doi: 10.7554/eLife.23485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta A, Thiruvengadam G, Desai SA. 2015. The conserved clag multigene family of malaria parasites: essential roles in host-pathogen interaction. Drug Resist Update 18:47–54. doi: 10.1016/j.drup.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling IT, Kaneko O, Narum DL, Tsuboi T, Howell S, Taylor HM, Scott-Finnigan TJ, Torii M, Holder AA. 2003. Characterization of the rhoph2 gene of Plasmodium falciparum and Plasmodium yoelii. Mol Biochem Parasitol 127:47–57. doi: 10.1016/s0166-6851(02)00302-x. [DOI] [PubMed] [Google Scholar]
- 8.Shirano M, Tsuboi T, Kaneko O, Tachibana M, Adams JH, Torii M. 2001. Conserved regions of the Plasmodium yoelii rhoptry protein RhopH3 revealed by comparison with the P falciparum homologue. Mol Biochem Parasitol 112:297–299. doi: 10.1016/S0166-6851(00)00366-2. [DOI] [PubMed] [Google Scholar]
- 9.Holt DC, Fischer K, Tchavtchitch M, Wilson DW, Hauquitz NE, Hawthorne PL, Gardiner DL, Trenholme KR, Kemp DJ. 2001. Clags in Plasmodium falciparum and other species of Plasmodium. Mol Biochem Parasitol 118:259–263. doi: 10.1016/s0166-6851(01)00378-4. [DOI] [PubMed] [Google Scholar]
- 10.Otto TD, Bohme U, Sanders M, Reid A, Bruske EI, Duffy CW, Bull PC, Pearson RD, Abdi A, Dimonte S, Stewart LB, Campino S, Kekre M, Hamilton WL, Claessens A, Volkman SK, Ndiaye D, Amambua-Ngwa A, Diakite M, Fairhurst RM, Conway DJ, Franck M, Newbold CI, Berriman M. 2018. Long read assemblies of geographically dispersed Plasmodium falciparum isolates reveal highly structured subtelomeres. Wellcome Open Res 3:52. doi: 10.12688/wellcomeopenres.14571.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cortes A, Carret C, Kaneko O, Yim Lim BY, Ivens A, Holder AA. 2007. Epigenetic silencing of Plasmodium falciparum genes linked to erythrocyte invasion. PLoS Pathog 3:e107. doi: 10.1371/journal.ppat.0030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pillai AD, Nguitragool W, Lyko B, Dolinta K, Butler MM, Nguyen ST, Peet NP, Bowlin TL, Desai SA. 2012. Solute restriction reveals an essential role for clag3-associated channels in malaria parasite nutrient acquisition. Mol Pharmacol 82:1104–1114. doi: 10.1124/mol.112.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma P, Wollenberg K, Sellers M, Zainabadi K, Galinsky K, Moss E, Nguitragool W, Neafsey D, Desai SA. 2013. An epigenetic antimalarial resistance mechanism involving parasite genes linked to nutrient uptake. J Biol Chem 288:19429–19440. doi: 10.1074/jbc.M113.468371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguitragool W, Rayavara K, Desai SA. 2014. Proteolysis at a specific extracellular residue implicates integral membrane CLAG3 in malaria parasite nutrient channels. PLoS One 9:e93759. doi: 10.1371/journal.pone.0093759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rovira-Graells N, Crowley VM, Bancells C, Mira-Martinez S, Ribas de PL, Cortes A. 2015. Deciphering the principles that govern mutually exclusive expression of Plasmodium falciparum clag3 genes. Nucleic Acids Res 43:8243–8257. doi: 10.1093/nar/gkv730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mira-Martinez S, Pickford AK, Rovira-Graells N, Guetens P, Tinto-Font E, Cortes A, Rosanas-Urgell A. 2019. Identification of antimalarial compounds that require CLAG3 for their uptake by Plasmodium falciparum-infected erythrocytes. Antimicrob Agents Chemother 63:e00052-19. doi: 10.1128/AAC.00052-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coppel RL, Bianco AE, Culvenor JG, Crewther PE, Brown GV, Anders RF, Kemp DJ. 1987. A cDNA clone expressing a rhoptry protein of Plasmodium falciparum. Mol Biochem Parasitol 25:73–81. doi: 10.1016/0166-6851(87)90020-x. [DOI] [PubMed] [Google Scholar]
- 18.Ling IT, Florens L, Dluzewski AR, Kaneko O, Grainger M, Yim Lim BY, Tsuboi T, Hopkins JM, Johnson JR, Torii M, Bannister LH, Yates JR III, Holder AA, Mattei D. 2004. The Plasmodium falciparum clag9 gene encodes a rhoptry protein that is transferred to the host erythrocyte upon invasion. Mol Microbiol 52:107–118. doi: 10.1111/j.1365-2958.2003.03969.x. [DOI] [PubMed] [Google Scholar]
- 19.Ghoneim A, Kaneko O, Tsuboi T, Torii M. 2007. The Plasmodium falciparum RhopH2 promoter and first 24 amino acids are sufficient to target proteins to the rhoptries. Parasitol Int 56:31–43. doi: 10.1016/j.parint.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Vincensini L, Fall G, Berry L, Blisnick T, Braun BC. 2008. The RhopH complex is transferred to the host cell cytoplasm following red blood cell invasion by Plasmodium falciparum. Mol Biochem Parasitol 160:81–89. doi: 10.1016/j.molbiopara.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Shaner NC, Steinbach PA, Tsien RY. 2005. A guide to choosing fluorescent proteins. Nat Methods 2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 22.Soleja N, Manzoor O, Khan I, Ahmad A, Mohsin M. 2018. Role of green fluorescent proteins and their variants in development of FRET-based sensors. J Biosci 43:763–784. doi: 10.1007/s12038-018-9783-0. [DOI] [PubMed] [Google Scholar]
- 23.Piston DW, Kremers GJ. 2007. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem Sci 32:407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Iriko H, Kaneko O, Otsuki H, Tsuboi T, Su XZ, Tanabe K, Torii M. 2008. Diversity and evolution of the rhoph1/clag multigene family of Plasmodium falciparum. Mol Biochem Parasitol 158:11–21. doi: 10.1016/j.molbiopara.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta A, Balabaskaran-Nina P, Nguitragool W, Saggu GS, Schureck MA, Desai SA. 2018. CLAG3 self-associates in malaria parasites and quantitatively determines nutrient uptake channels at the host membrane. mBio 9:e02293-17. doi: 10.1128/mBio.02293-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beck JR, Muralidharan V, Oksman A, Goldberg DE. 2014. PTEX component HSP101 mediates export of diverse malaria effectors into host erythrocytes. Nature 511:592–595. doi: 10.1038/nature13574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desai SA, Bezrukov SM, Zimmerberg J. 2000. A voltage-dependent channel involved in nutrient uptake by red blood cells infected with the malaria parasite. Nature 406:1001–1005. doi: 10.1038/35023000. [DOI] [PubMed] [Google Scholar]
- 28.Kenworthy AK. 2001. Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods 24:289–296. doi: 10.1006/meth.2001.1189. [DOI] [PubMed] [Google Scholar]
- 29.Jares-Erijman EA, Jovin TM. 2006. Imaging molecular interactions in living cells by FRET microscopy. Curr Opin Chem Biol 10:409–416. doi: 10.1016/j.cbpa.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 30.Wallrabe H, Barroso M. 2005. Confocal FRET microscopy: study of clustered distribution of receptor-ligand complexes in endocytic membranes, p 95–111. In Periasamy A, Day RN (ed), Molecular imaging: FRET microscopy and spectroscopy. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 31.Koushik SV, Chen H, Thaler C, Puhl HL III, Vogel SS. 2006. Cerulean, Venus, and VenusY67C FRET reference standards. Biophys J 91:L99–L101. doi: 10.1529/biophysj.106.096206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Y, Day RN, Periasamy A. 2011. Investigating protein-protein interactions in living cells using fluorescence lifetime imaging microscopy. Nat Protoc 6:1324–1340. doi: 10.1038/nprot.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maier AG, Cooke BM, Cowman AF, Tilley L. 2009. Malaria parasite proteins that remodel the host erythrocyte. Nat Rev Microbiol 7:341–354. doi: 10.1038/nrmicro2110. [DOI] [PubMed] [Google Scholar]
- 34.Spielmann T, Gilberger TW. 2015. Critical steps in protein export of Plasmodium falciparum blood stages. Trends Parasitol 31:514–525. doi: 10.1016/j.pt.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 35.Desai SA. 2012. Ion and nutrient uptake by malaria parasite-infected erythrocytes. Cell Microbiol 14:1003–1009. doi: 10.1111/j.1462-5822.2012.01790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell GH, Miller LH, Hudson D, Franco EL, Andrysiak PM. 1984. Monoclonal antibody characterization of Plasmodium falciparum antigens. Am J Trop Med Hyg 33:1051–1054. doi: 10.4269/ajtmh.1984.33.1051. [DOI] [PubMed] [Google Scholar]
- 37.Holder AA, Freeman RR, Uni S, Aikawa M. 1985. Isolation of a Plasmodium falciparum rhoptry protein. Mol Biochem Parasitol 14:293–303. doi: 10.1016/0166-6851(85)90057-x. [DOI] [PubMed] [Google Scholar]
- 38.Kaneko O, Yim Lim BY, Iriko H, Ling IT, Otsuki H, Grainger M, Tsuboi T, Adams JH, Mattei D, Holder AA, Torii M. 2005. Apical expression of three RhopH1/Clag proteins as components of the Plasmodium falciparum RhopH complex. Mol Biochem Parasitol 143:20–28. doi: 10.1016/j.molbiopara.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 39.Gardiner DL, Skinner-Adams TS, Spielmann T, Trenholme KR. 2003. Malaria transfection and transfection vectors. Trends Parasitol 19:381–383. doi: 10.1016/S1471-4922(03)00187-9. [DOI] [PubMed] [Google Scholar]
- 40.Limenitakis J, Soldati-Favre D. 2011. Functional genetics in Apicomplexa: potentials and limits. FEBS Lett 585:1579–1588. doi: 10.1016/j.febslet.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 41.Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio JJ. 2014. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol 32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 42.Esposito A, Tiffert T, Mauritz JM, Schlachter S, Bannister LH, Kaminski CF, Lew VL. 2008. FRET imaging of hemoglobin concentration in Plasmodium falciparum-infected red cells. PLoS One 3:e3780. doi: 10.1371/journal.pone.0003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kraft TE, Heitmeier MR, Putanko M, Edwards RL, Ilagan MX, Payne MA, Autry JM, Thomas DD, Odom AR, Hruz PW. 2016. A novel fluorescence resonance energy transfer-based screen in high-throughput format to identify inhibitors of malarial and human glucose transporters. Antimicrob Agents Chemother 60:7407–7414. doi: 10.1128/AAC.00218-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abshire JR, Rowlands CJ, Ganesan SM, So PT, Niles JC. 2017. Quantification of labile heme in live malaria parasites using a genetically encoded biosensor. Proc Natl Acad Sci U S A 114:E2068–E2076. doi: 10.1073/pnas.1615195114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Topolska AE, Lidgett A, Truman D, Fujioka H, Coppel RL. 2004. Characterization of a membrane-associated rhoptry protein of Plasmodium falciparum. J Biol Chem 279:4648–4656. doi: 10.1074/jbc.M307859200. [DOI] [PubMed] [Google Scholar]
- 46.Gunalan K, Gao X, Yap SSL, Lai SK, Ravasio A, Ganesan S, Li HY, Preiser PR. 2020. A processing product of the Plasmodium falciparum reticulocyte binding protein RH1 shows a close association with AMA1 during junction formation. Cell Microbiol 2020:e13232. [DOI] [PubMed] [Google Scholar]
- 47.Fung BK, Stryer L. 1978. Surface density determination in membranes by fluorescence energy transfer. Biochemistry 17:5241–5248. doi: 10.1021/bi00617a025. [DOI] [PubMed] [Google Scholar]
- 48.Vogel SS, Thaler C, Koushik SV. 2006. Fanciful FRET. Sci STKE 2006:re2. doi: 10.1126/stke.3312006re2. [DOI] [PubMed] [Google Scholar]
- 49.Broussard JA, Rappaz B, Webb DJ, Brown CM. 2013. Fluorescence resonance energy transfer microscopy as demonstrated by measuring the activation of the serine/threonine kinase Akt. Nat Protoc 8:265–281. doi: 10.1038/nprot.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomavo S, Slomianny C, Meissner M, Carruthers VB. 2013. Protein trafficking through the endosomal system prepares intracellular parasites for a home invasion. PLoS Pathog 9:e1003629. doi: 10.1371/journal.ppat.1003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cobb DW, Florentin A, Fierro MA, Krakowiak M, Moore JM, Muralidharan V. 2017. The exported chaperone PfHsp70x is dispensable for the Plasmodium falciparum intraerythrocytic life cycle. mSphere 2:e00363-17. doi: 10.1128/mSphere.00363-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McHugh E, Carmo OMS, Blanch A, Looker O, Liu B, Tiash S, Andrew D, Batinovic S, Low AJY, Cho HJ, McMillan P, Tilley L, Dixon MWA. 2020. Role of Plasmodium falciparum protein GEXP07 in Maurer’s cleft morphology, knob architecture, and P falciparum EMP1 trafficking. mBio 11:e03320-19. doi: 10.1128/mBio.03320-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta A, Bokhari AA, Pillai AD, Crater AK, Gezelle J, Saggu GS, Nasamu AS, Ganesan SM, Niles JC, Desai SA. 2020. Complex nutrient channel phenotypes despite Mendelian inheritance in a P falciparum genetic cross. PLoS Pathog 16:e1008363. doi: 10.1371/journal.ppat.1008363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cohn JV, Alkhalil A, Wagner MA, Rajapandi T, Desai SA. 2003. Extracellular lysines on the plasmodial surface anion channel involved in Na+ exclusion. Mol Biochem Parasitol 132:27–34. doi: 10.1016/j.molbiopara.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 55.Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. 2014. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ribeiro JM, Garriga M, Potchen N, Crater AK, Gupta A, Ito D, Desai SA. 2018. Guide RNA selection for CRISPR-Cas9 transfections in Plasmodium falciparum. Int J Parasitol 48:825–832. doi: 10.1016/j.ijpara.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Production of R2mV and C3mC parasites. (A) CRISPR/Cas9 strategy for tagging of the endogenous rhoph2 gene with mVenus. Site-specific cleavage of rhoph2 by the Cas9-sgRNA complex is repaired through homology-directed repair with homology arms that flank an in-frame mVenus open reading frame. (B) Ethidium stained gels showing PCR integration checks for R2mV and its wild-type KC5 parent. The R2mV clone has fully replaced the wild-type locus with the desired mVenus tag. (C) Full-length R2mV immunoblot, showing the entire blot from Fig. 1B. (D) Strategy for clag3 tagging with mCerulean. (D) Ethidium-stained gels showing PCR integration checks for C3mC and its KC5 parent. Primer positions are as indicated in panel A; primer sequences are provided in Table S1 in the supplemental material. (F) Full-length C3mC immunoblot from Fig. 1B. Download FIG S1, PDF file, 0.3 MB (347.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Sequential transfections to produce double-tagged parasites. (A) Flowchart showing strategy for production of C3R2-DKI. (B) Ethidium-stained gels showing PCR integration checks for C3R2-DKI and its KC5 parent. (C) Full-length immunoblot from Fig. 3B. (D) Strategy for production of R2mV-LmC. (E) PCR confirming preserved integrated of the mVenus tag on rhoph2 and retention of the episome for expression of mCerulean with a RhopH2 leader sequence in R2mV-LmC parasites. Download FIG S2, PDF file, 0.3 MB (298.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
R3-C5V FRET positive-control parasite. (A) CRISPR/Cas9 strategy. (B) PCR integration checks for the R3-C5V clone and its wild-type KC5 parent. Primer sequences are in Table S1 in the supplemental material. (C) Immunoblots probed with anti-GFP showing absence of signal in untransfected wild-type cells (WT) and the tandem mCerulean-mVenus tagged RhopH3 protein (band at 150 kDa) in R3-C5V parasites. The blot on the right shows a stage-specific cleavage product at 45 kDa that results from proteolytic processing of RhopH3 in parasite rhoptries (5). Download FIG S3, PDF file, 0.3 MB (366.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Primers and sgRNAs used in this study. Download Table S1, XLSX file, 0.01 MB (12.3KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.