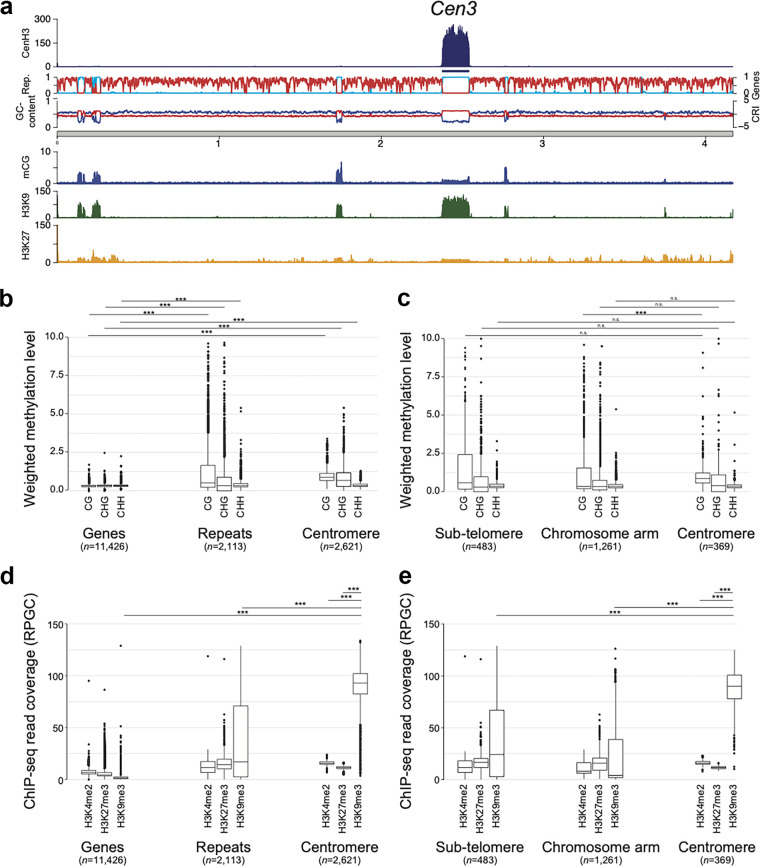
FIG 2.
Centromeres in Verticillium dahliae strain JR2 are embedded in heterochromatin. (a) Schematic overview of chromosome 3 of V. dahliae strain JR2, exemplifying the distribution of heterochromatin-associated chromatin modifications (mC, H3K9me3, and H3K27me3) in relation to the centromeres. The different lanes display the FLAG-CenH3 ChIP-seq read coverage (RPGC normalization in 1-kb bins with 3-kb smoothening), the FLAG-CenH3 enriched regions, the repeat and gene density (light blue and red, respectively), the GC content (blue), the CRI (red) as well as the weighted cytosine methylation (all summarized in 5-kb windows with 500-bp slide), and the normalized H3K9me3 and H3K27me3 ChIP-seq read coverage (RPGC normalization in 1-kb bins with 3-kb smoothening). The schematic overview of all chromosomes is shown in Fig. S2. (b) Boxplots of weighted DNA methylation levels per genomic context (CG, CHG, or CHH) are summarized over genes, repetitive elements, or 5-kb genomic windows (500-bp slide) overlapping the centromeric regions. (c) Weighted DNA methylation levels per genomic context (CG, CHG, or CHH) are summarized over repetitive elements that have been split based on their genomic location: subtelomeres (within the first or last 10% of the chromosome), centromeres, or the remainder of the chromosome arm. (d) ChIP-seq read coverage (RPGC normalized; see panel a for H3K4me2, H3K27m3, and H3K9me3) is summarized over genes, repetitive elements, or 5-kb windows (500-bp slide) overlapping the centromeric regions. (e) ChIP-seq read coverage (RPGC normalized; see panel a for H3K4me2, H3K27m3, and H3K9me3) is summarized over repetitive elements that have been split based on their genomic location: subtelomeres (within the first or last 10% of the chromosome), centromeres, or the remainder of the chromosomal arm. Statistical differences for the indicated comparisons were calculated using the one-sided nonparametric Mann-Whitney test; P values <0.001, ***; n.s., not significant.