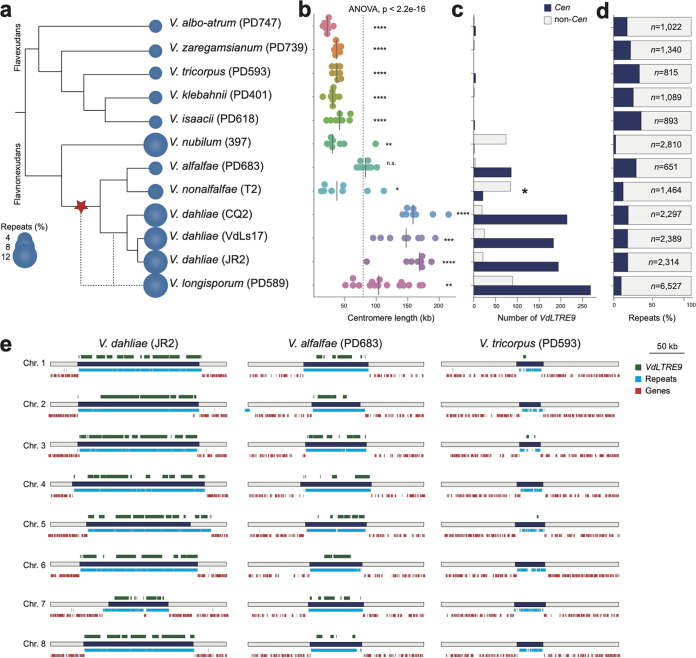
FIG 5.
Evolution of centromeres in the genus Verticillium. (a) Relationship of the 10 members of the genus Verticillium. The predicted repeat content for each of the genomes is indicated (see Table S1c for details). The red star indicates the recruitment of VdLTRE9 into centromeres. (b) Comparison of estimated centromere lengths (in kb) in the different Verticillium spp. Each dot represents a single centromere, and the line represents the median size. (c) The number of (partial) VdLTRE9 matches identified in centromeres (Cen; dark blue) and across the genome (non-Cen; light gray). The asterisk indicates the high number of VdLTRE9 elements in unassigned contigs for Verticillium nonalfalfae strain T2 (see the text for details). (d) Proportion of predicted repeat content localized at centromeres (Cen; dark blue) and across the genome (non-Cen; light gray). (e) Schematic overview of the eight centromeric regions (250 kb) in Verticillium dahliae strain JR2, and Verticillium alfalfae strain PD683 and Verticillium tricorpus strain PD593 as representatives for clade Flavnonexudans and clade Flavexudans, respectively. The centromeres are indicated by dark blue bars. The predicted genes (red) and repeats (light blue) are shown below each centromere, and locations of (partial) VdLTRE9 matches (light green) are shown above each centromere. Global statistical differences for the centromere sizes were calculated using one-way analysis of variance (ANOVA), and differences for each species compared to the overall mean were computed using unpaired t tests; P values <0.0001, ****; P values <0.001, ***; P values <0.01, **; P values <0.05, *; n.s., not significant.