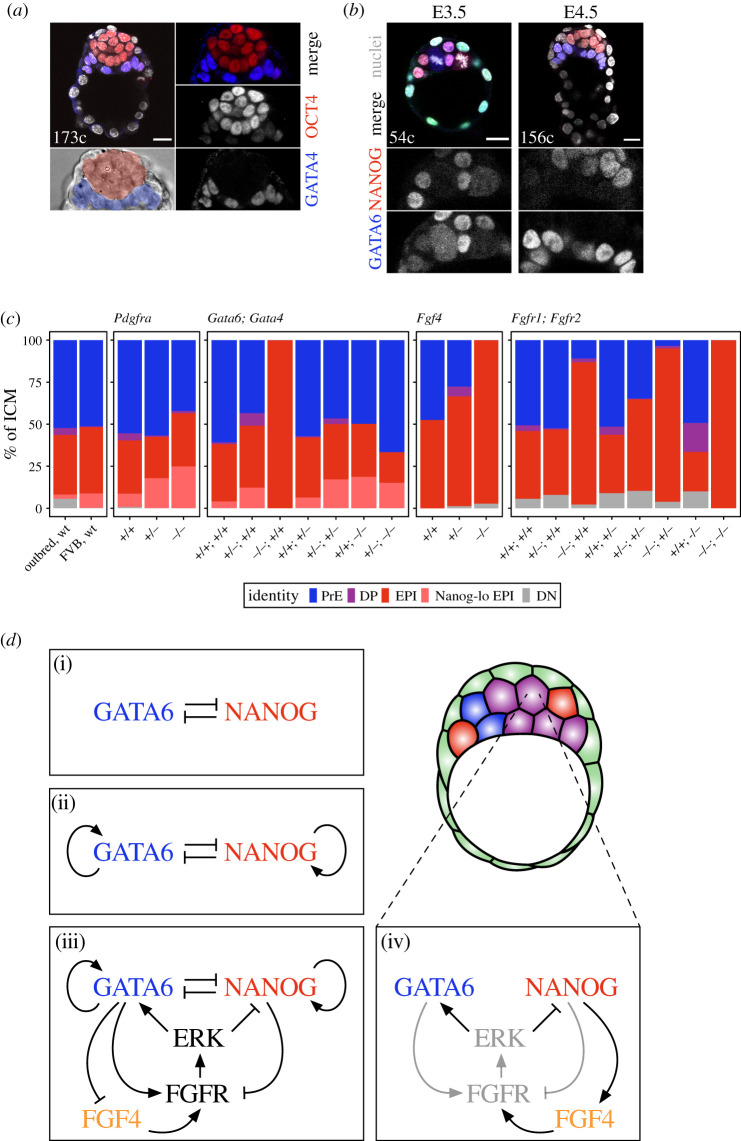
Figure 3.
Ways to achieve a balanced lineage composition in the ICM. (a) Immunofluorescence images of an E4.5 blastocyst with sorted epiblast (marked by OCT4, red) and PrE (marked by GATA4, blue). At this stage, the PrE forms a coherent epithelial layer between the epiblast and the blastocyst cavity. (b) Immunofluorescence images of an E3.5 and an E4.5 embryo stained for NANOG (epiblast marker, red) and GATA6 (PrE marker, blue). At E3.5, all or most ICM cells co-express both NANOG and GATA6. At E4.5, the two markers label the sorted populations, as shown in (a). (c) Stacked bar plots showing ICM composition in late blastocysts (greater than 100 cells) of the genotypes indicated. Composition for embryos from an outbred (CD1) and an inbred (FVB) line is shown in the first box. (d) Different theoretical models to explain lineage specification within the ICM: direct mutual inhibition between GATA6 and NANOG (i), direct mutual inhibition with auto-activation (ii), direct mutual inhibition with auto-activation and modulation by FGF–MAPK signalling (iii) and indirect mutual inhibition via FGF–MAPK signalling (iv). Black arrows in (iv) represent interactions explicitly modelled in [119], whereas grey arrows represent interactions that are implicit in that model. Note how the net effect of these interactions is equivalent to the model shown in (ii) (e.g. GATA6 inhibits NANOG via the activation of FGFR, not directly). See the text for details. Colour coding is indicated. EPI, epiblast; Nanog-lo EPI, NANOG-low epiblast; PrE, primitive endoderm; DP, double-positive (for NANOG and GATA6); DN, double negative (for NANOG and GATA6). Scale bar, 20 µm.