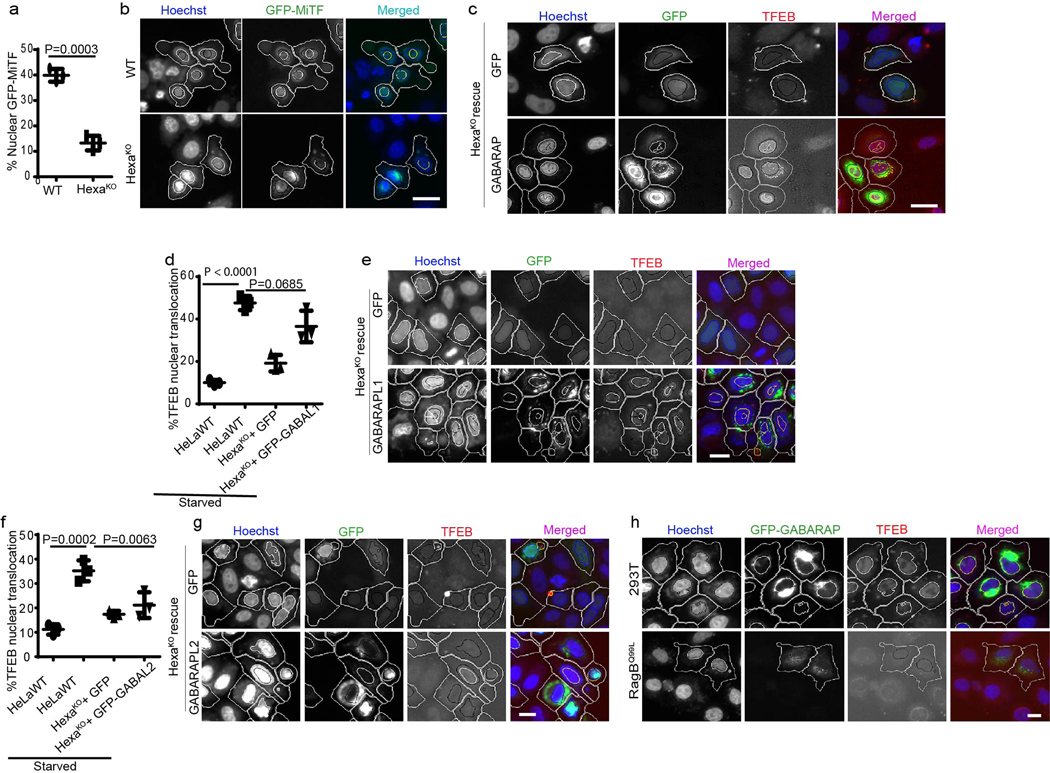
Extended Data Fig. 4. GABARAP and GABARAPL1 but not GABARAPL2 control nuclear translocation of TFEB.
a,b, HCM images and quantifications to test the role of mAtg8s on nuclear translocation of GFP-MiTF in response to autophagy induction (EBSS 2h). Data, means ± SEM (n=3 biologically independent experiments) paired t-test. Masks; white: algorithm-defined cell boundaries; blue outline: computer-identified nuclear stain; yellow outline: computer-identified colocalization between TFEB and Hoechst-33342 nuclear stain). Scale bar 10 μm. The masks in gray scale panels are cloned from the merged images, (n=3 biologically independent experiments). c, HCM image analysis of effects of complementation of HexaKO with GFP-GABARAP on nuclear translocation of TFEB. Scale bar 10 μm. d–g, HCM analysis of the effect of complementation of HexaKO cells with GABARAPL1 or GABARAPL2 on nuclear translocation of TFEB. Data, means ± SEM, ANOVA, Tukey’s post hoc test; HCM, >500 cells counted per well; minimum number of valid wells 9, (n=3 biologically independent experiments). Scale bar 10 μm. h, HCM analysis of effect of expression of GABARAP in 293T cells expressing RagBQ99L or parental 293T cells on nuclear translocation of TFEB. Masks in c, e, g, h; white: algorithm-defined cell boundaries in GFP positive cells; blue outline: computer-identified nuclear stain; yellow outline: computer-identified co-localization between TFEB and Hoechst-33342 nuclear stain), (n=3 biologically independent experiments). Scale bar 10 μm. Numerical source data for panels a, d, and f are provided in Statistical Source Data Extended Fig. 4.