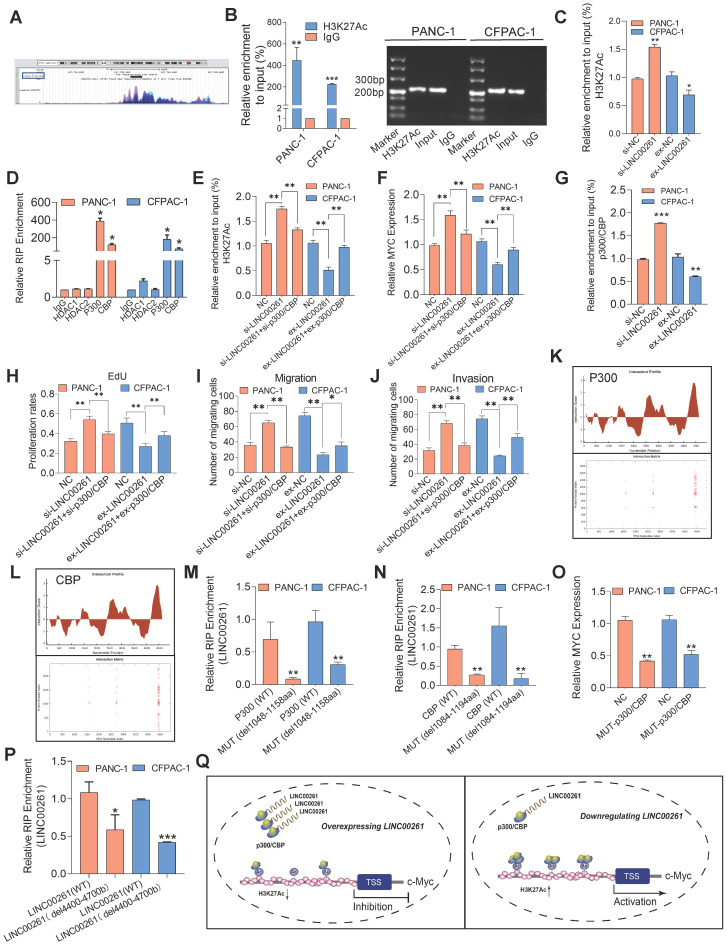
Figure 7.
LINC00261 inhibits c-Myc-driven malignant phenotypes via p300/CBP. (A) H3K27 acetylation of the c-Myc promoter region in the UCSC genome database. (B) ChIP assays with an anti-H3K27Ac antibody or IgG were performed to verify the enrichment of H3K27Ac at binding sites in the c-Myc promoter in PC cells. The ChIP-PCR products in the c-Myc, input and IgG groups were evaluated by agarose gel electrophoresis. (C) ChIP assays with an anti-H3K27Ac antibody or IgG were performed in PC cells with LINC00261 knockdown, PC cells with LINC00261 overexpression and control cells. (D) RIP assays were performed to validate LINC00261 binding to related proteins in PANC-1 and CFPAC-1 cells. (E) ChIP assays with an anti-H3K27Ac antibody or IgG were performed in the three groups of PANC-1 cells (si-NC, si-LINC00261, si-LINC00261+si-p300/CBP) and in the three groups of CFPAC-1 cells (ex-NC, ex-LINC00261 and ex-LINC00261+ex-p300/CBP). (F) c-Myc gene expression levels were analyzed by qRT-PCR in the three groups of PANC-1 cells (si-NC, si-LINC00261, si-LINC00261+si-p300/CBP) and in the three groups of CFPAC-1 cells (ex-NC, ex-LINC00261 and ex-LINC00261+ex-p300/CBP). (G) ChIP assays with an anti-p300/CBP antibody or IgG were performed to verify the enrichment of p300/CBP at binding sites in the c-Myc promoter in PC cells with LINC00261 downregulation and overexpression. (H) Histogram showing the proliferation rates of cotransfected PC cells. (I and J) Histograms showing the number of migrated and invaded cotransfected PC cells. (K and L) The interaction profile, which represents the P300/CBP interaction score (Y-axis) relative to the LINC00261 RNA sequence (X-axis), provides information about the region most likely to be bound by the protein (up); The interaction matrix, which shows a heatmap of the P300/CBP(Y-axis) and LINC00261 RNA (X-axis) regions. The red shading in the heatmap indicates the interaction score of a single amino acid and nucleotide pair (down). (M and N) RIP assays with an anti-Flag antibody or IgG were performed to verify the LINC00261 enrichment in PANC-1 and CFPAC-1 cells transfected with the p300 vector (WT) or mutant p300 vector (del-1048-1158aa) and the CBP vector (WT) or the mutant CBP vector (del-1084-1194aa). All plasmids were labeled with 3-flag. (O) The mRNA levels of c-Myc was assessed by qRT-PCR, in the NC and mut-p300/CBP groups. (P) RIP assays were performed to validate p300/CBP binding sites of LINC00261 region in PANC-1 and CFPAC-1 cells transfected with LINC00261 (WT) and mutant LINC00261 vector (del-4400bp-4700bp). (Q) A schematic model of the mechanism underlying the role of LINC00261 in PC progression (*P < 0.05, **P < 0.01, ***P < 0.001).