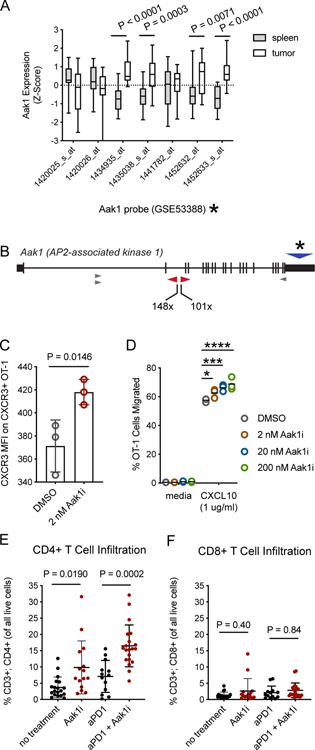
Figure 4. Inhibition of Aak1 kinase activity enhances mouse T-cell migration.
A, Publicly available microarray data (7) was used to evaluate Aak1 gene expression of intratumoral T cells compared to splenic T cells from the same mice (n = 3 mice). Probe data for Aak1 was pulled and differential expression P values were determined by two-sided t-test for individual probes. All probes hybridized to exon 21 of Aak1. Bars indicate population mean, standard deviation, and min/max. Asterisk refers to probe location in panel B. B, Aak1 was the most frequently mutated gene in our screen. Transposon insertions were predominately clustered in intron 2, as indicated by the red triangles, with a few other insertion sites elsewhere in the gene (grey triangles). Though insertions were tightly clustered, transposon orientation was unbiased. Asterisk and blue triangle represent the region covered by microarray probes panel A. C, Cell surface expression of Cxcr3 on activated, primary mouse T cells treated with Aak1i or DMSO control for 1.5 hours was measured by flow cytometry. Each dot represents a technical replicate, and data are representative of three biological replicates (one-way ANOVA, error bars represent standard deviation, experiment was replicated three times). D, Migration of primary mouse T cells toward CXCL10 with Aak1i or DMSO vehicle control treatment (*P=0.03, ***P=0.0004, ****P<0.0001, two-way ANOVA with multiple comparisons, horizontal lines indicate means). E,F, B16F0 melanoma tumors treated with Aak1i or DMSO vehicle control were harvested at tumor growth day 21, and (E) CD4+ and (F) CD8+ T-cell infiltration was measured by flow cytometry. Each dot represents an individual mouse. Significance determined by one-way ANOVA. Data shown are representative of 3 replicates with 5 mice/group in each replicate.