Abstract
The HIV-1 Nef protein suppresses multiple immune surveillance mechanisms to promote viral pathogenesis and is an attractive target for the development of novel therapeutics. A key function of Nef is to remove the CD4 receptor from the cell surface by hijacking clathrin- and AP2-dependent endocytosis. However, exactly how Nef does this has been elusive. Here, we describe the underlying mechanism as revealed by a 3.0Å crystal structure of a fusion protein comprised of Nef and the cytoplasmic domain of CD4 bound to the tetrameric AP2 complex. An intricate combination of conformational changes occurs in both Nef and AP2 to enable CD4 binding and downregulation. A pocket on Nef previously identified as crucial for recruiting class I MHC is also responsible for recruiting CD4, revealing a potential approach to inhibit two of Nef’s activities and sensitize the virus to immune clearance.
Introduction
The HIV-1 protein Nef is a critical factor in viral pathogenesis1. Expression of Nef in vivo is required for high viral loads and for progression to AIDS2. Individuals infected with HIV-1 encoding defective nef genes do not develop AIDS for decades3,4. Nef is a peripheral membrane protein that anchors to the lipid membrane via a N-terminal myristoyl group. Abundantly expressed early during the viral replication cycle, Nef modulates the surface levels of many host proteins and thereby interferes with immune processes responsible for detecting and combating the infection1,5,6. Two of the most prominent targets of Nef are MHC-I, which is crucial for antigen presentation and the killing of infected cells by cytotoxic T lymphocytes, and CD4.
CD4 is the entry receptor for HIV-1, but later in the viral replication cycle it is problematic for the virus: it disrupts processing of the viral glycoprotein, Env, inhibiting infectivity7; it interferes with the release of new virions8,9; and it causes vulnerability to superinfection, causing premature cell death and limiting viral productivity10. Furthermore, binding of CD4 to Env exposes otherwise-concealed Env epitopes, rendering infected cells more susceptible to antibody-dependent cellular cytotoxicity and virus particles more susceptible to neutralizing antibodies11–13. HIV-1 has evolved strategies to mitigate these problems. Newly synthesized CD4 is targeted in the ER by the viral Vpu protein for proteasomal degradation14. Surface-expressed CD4, in contrast, is targeted by Nef for endocytosis and lysosomal degradation15–18.
Nef’s effect on CD4 involves hijacking of clathrin adaptor protein complex 2 (AP2)-dependent endocytosis19,20. Adaptor protein complexes mediate the clathrin-dependent trafficking of membrane proteins21. Members of this family (AP1–5) share a common heterotetrameric structure, which adopts either a closed or an open conformation. When activated and open, AP complexes reveal their cargo-bindings sites, which then associate with specific sorting motifs in the cytoplasmic domains of membrane cargos. Two sorting motifs are commonly recognized by APs. The tyrosine-based motifs (YxxΦ where Φ is a bulky hydrophobic residue) bind to the μ subunits of APs, while acidic dileucine motifs ([E/D]xxxL[L/I]) bind to a pocket formed largely by the σ subunits. To mediate downregulation of CD4 from the cell surface, Nef co-opts the clathrin AP2 complex partly by mimicking the acidic dileucine motif22. Although how Nef associates with a part of the tetrameric AP2 is understood22, a complete understanding of the interaction, especially how CD4 is sequestered by Nef into a complex with AP2, has remained elusive. Here we describe the mechanism of this key Nef function using a high-resolution crystal structure, cross-linking mass spectrometry, and structure-modeling, along with biochemical and functional validation.
Results
Nef functions as a “connector” between clathrin AP2 and CD4.
We pursued a high-resolution structure of Nef in complex with tetrameric AP2 and the cytoplasmic domain of CD4 (CD4CD). To facilitate the assembly of the protein complex, we fused CD4CD to the C-terminus of Nef via a 36 amino acid-long, flexible linker (Fig. 1a). Instead of using the full length CD4 tail (394–433), we included only residues 394 to 419, containing all the CD4 determinants reportedly required for Nef-mediated downregulation16,23,24. Since Nef residues within the N-terminal amphipathic helix are dispensable for CD4 downregulation25, we truncated 25 amino acids from the Nef N-terminus. We engineered the tetrameric AP2 complex by removing the mobile C-terminal domain of the μ2 subunit (136–423), enabling AP2 to adopt an open conformation in which its cargo-binding sites are accessible26. Binding between the Nef-CD4CD fusion protein and the modified AP2 construct (AP2Δμ2-CTD) was confirmed using a GST pulldown assay (Fig. 1b).
Fig. 1. In vitro assembly and crystal structure of the Nef, CD4 cytoplasmic domain, and clathrin AP2 complex.
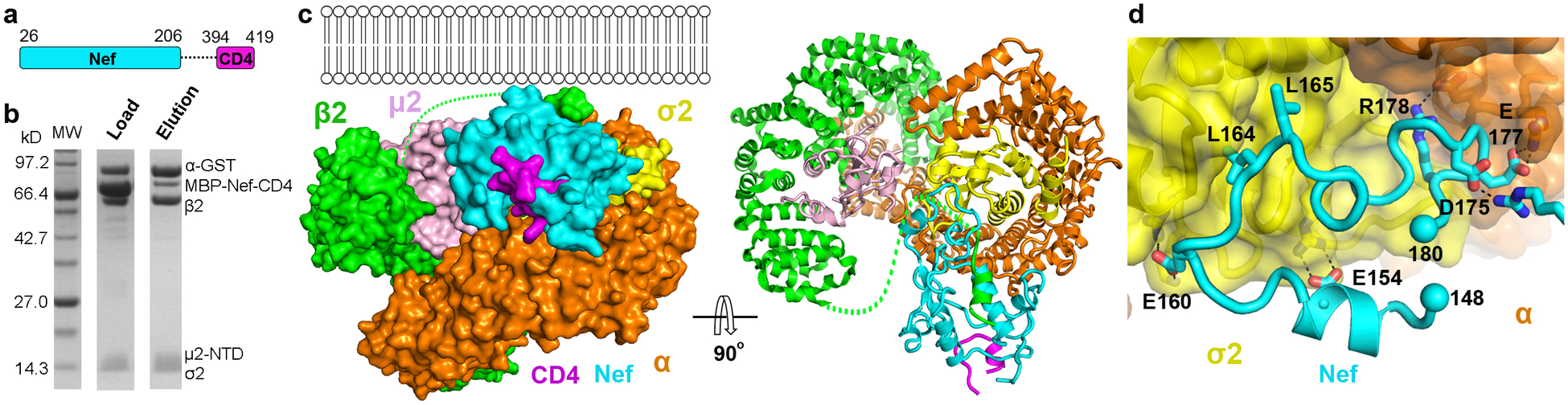
a, Cartoon illustrating the design of the Nef-CD4CD fusion protein. b, In vitro GST pulldown assay confirming the binding between the MBP-Nef-CD4CD fusion protein and the AP2Δμ2-CTD complex. c, Crystal structure of the complete protein complex in two views: along the membrane plane (left) and downward from the membrane (right). d, Nef associates with AP2 mainly through its C-terminal loop (148–180). The rest of Nef is not shown for an unblocked view.
We then solved the crystal structure of the Nef-CD4CD fusion and AP2Δμ2-CTD complex to a resolution of 3.0Å (Fig. 1c, Table 1). All polypeptides are largely resolved except for the flexible linker between Nef and CD4CD, which is disordered as expected, and part of the N-terminal region of β2. As revealed by the structure, Nef functions as a “connector” between AP2 and CD4CD; CD4CD binds Nef but none of the subunits of AP2. As previously reported, Nef’s C-terminal loop interacts with AP2 in part via mimicry of the acidic dileucine motif (Fig. 1d)22. The extensive interface here, involving several charge-charge and hydrophobic interactions, is the foundation of the Nef-AP2 association.
Table 1.
Crystallographic data collection and refinement statistics
| Nef-CD4CD–AP2Δμ2-CTD (PDB 6URI) | |
|---|---|
| Data collection | |
| Space group | P41 |
| Cell dimensions | |
| a, b, c (Å) | 109.2, 109.2, 178.9 |
| Wavelength (Å) | 0.9793 |
| Resolution (Å) | 47.2 – 3.0 (3.11 – 3.0) |
| Rmerge | 0.287 |
| I / σI | 11.8 (1.4) |
| Completeness (%) | 100 (100) |
| Redundancy | 16.0 (15.9) |
| Refinement | |
| Unique reflections | 41865 (4189) |
| Rwork / Rfree | 0.241/0.277 |
| No. atoms | |
| Protein | 12261 |
| Water | 2 |
| B-factors | |
| Protein | 118.30 |
| Water | 132.22 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 0.88 |
| Ramachandran | |
| Favored | 96.42% |
| Outliers | 0.13% |
Values in parenthesis are for the highest-resolution shell.
CD4 recruitment and role of the Nef N-terminal loop.
CD4 is recruited to a pocket on Nef that is opposite the C-terminal loop. The association is mainly hydrophobic and involves three CD4 residues: Ile410, Leu413, and Leu414 (Fig. 2a). The dileucine motif of CD4 - Leu413/414 - is within a short helix. Leu414 and Ile410 dip into a hydrophobic pocket of Nef, which is formed by Phe121, Trp124, Met79, Thr138, and Pro78 (Fig. 2b). Leu413 of CD4 sits just outside of that pocket and is accommodated by Nef residues Phe121, Leu37, Asn52, and Cys55 (Fig. 2b). Nef residue Asp123 contributes to CD4 binding by hydrogen-bonding with the backbone nitrogen of CD4 Leu413 and supports the helix-turn of CD4 (Fig. 2b). These structural findings explain previously observed roles of CD4 residues - the dileucine motif and Ile41016,24,27 - as well as Nef residues F121, D123, and Trp12428,29.
Fig. 2. Recruitment of CD4 cytoplasmic domain and the role of Nef N-terminal loop.
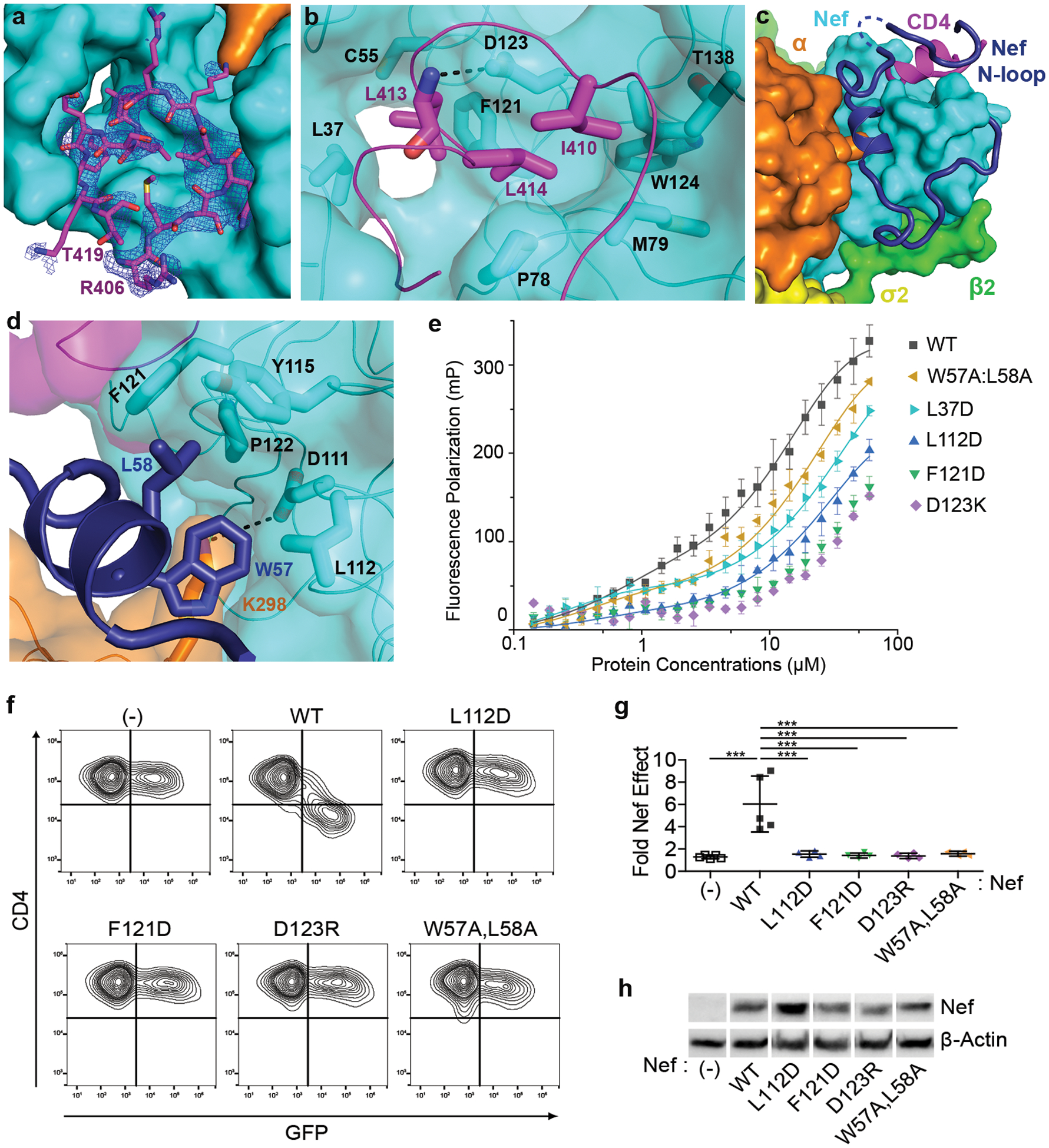
a, A short stretch of the CD4CD binds to a pocket on Nef. Electron density of the CD4CD (blue mesh, 2Fo-Fc map at 1.0σ with B factor sharpening of -50Å2) is shown. b, Hydrophobic interactions mediated by CD4 I410, L413, and L414. c, Nef N-terminal loop (dark blue) adopts a unique conformation in the complex. Part of the loop forms a wall of the CD4-binding pocket. Dotted line represents Nef 41–46 that are disordered in the structure. d, Nef residues W57 and L58 mediate the docking of the short helix into the hydrophobic pocket formed at the Nef-α interface. e, fluorescence polarization (FP) assay comparing WT and mutant Nef, when in complex with AP2, for their ability to bind tetramethylrhodamine(TMR)-CD4CD. FP is plotted as a function of Nef concentration (Nef:AP2=1:1.5, molar ratio). Data shown as mean and s.d. of three technical replicates. f, CD4 downregulation by Nef mutants was measured using cell surface-staining and flow cytometry. CD4-positive HeLa cells were transfected to express Nef and GFP; GFP is a transfection marker; GFP only control is indicated (-). “WT” indicates wild type Nef; Nef mutants are indicated. g, Quantification of the fold effect of Nef on the MFI of surface CD4, data shown as mean and s.d. for n = 4 (including L112D, F121D, D123R, W57A:L58A mutants) or n = 5 (including GFP only control, and WT Nef) independent transfections. Data were compared by one-way ANOVA with Bonferroni post test; ***p<0.0001. h, Expression of Nef proteins in the cells used in the CD4 downregulation assay measured by western blot; the cellular control is β-actin. Uncropped blot images are shown in Supplementary Figure 1. Data for graphs in e and g are available as source data.
CD4 recruitment is secured by the Nef N-terminal loop, which is highly ordered in the structure (Fig. 2c and Extended Data Fig. 1). The part of the loop immediately connected to the rigid core of Nef, residues Phe68 to Pro75, wraps around the core. N-terminally, the loop then takes a sharp “U-turn”, placing a short helix at the interface between the Nef core and the α subunit of AP2. Two residues within this helix, Trp57 and Leu58, fit into a hydrophobic pocket formed mainly by Nef residues: Leu112, Pro122, Phe121, and Tyr115 (Fig. 2d). Lys298 of α, with its head group stabilized by Nef Asp111 through charge-charge interactions, also contacts Nef Trp57 via its hydrocarbon chain (Fig. 2d). The Nef acidic cluster (Glu62–65), located in the “U-turn” region, does not contribute to CD4-binding, consistent with previous functional observations19,30,31. The Nef N-terminal loop extends to contact the CD4 tail (Fig. 2c). This part of Nef, residues Cys55 to Val33, forms a wall-like structure to support CD4-binding. Here, contacts are made by several Nef residues with the short helical turn of CD4 and flanking residues. Overall, this conformation of the Nef N-terminal loop explains the crucial roles of Trp57, Leu58, Leu112, Tyr115, and Pro122 in CD4-downregulation observed in previous studies19,20,25,28,32. It also explains the cooperativity in the three-way binding between CD4CD, Nef and the α/σ2 hemicomplex of AP233.
We then challenged these structural observations using mutagenesis. First, binding between a fluorescence tag-labeled CD4 cytoplasmic tail (tetramethylrhodamine-CD4CD; TMR-CD4CD) and the purified Nef/AP2 complex was characterized by an in vitro fluorescence polarization assay. While unbound TMR-CD4CD led to low polarization signal due to its fast tumbling and thus depolarization, binding of TMR-CD4CD to wild-type Nef and AP2 at higher protein concentrations resulted in drastic increase in molecular size, leading to slower tumbling and thereby high polarization signal (Fig. 2e). In contrast, reduced CD4-binding ability was observed for all Nef mutants tested. These mutations, according to the structure, affect either direct CD4 binding (D123K, L37D) or positioning of the Nef N-terminal short helix (W57A:L58A, L112D) or both (F121D). Second, these same Nef mutations were evaluated in a CD4 surface downregulation assay (Fig. 2f–h). HeLa cells that stably express CD4 were co-transfected with plasmids expressing GFP (as a transfection-marker) and plasmids expressing Nef. Two-color flow cytometry was used to measure cell-surface CD4 as a function of GFP-intensity. As expected, wild type Nef robustly downregulated CD4. Consistent with the in vitro fluorescence studies, CD4 downregulation was severely impaired for all the mutants tested (W57A:L58A, L112D, F121D, and D123R) (Fig. 2f–h). The L37D mutant was also inactive but poorly expressed (data not shown). Overall, these binding and functional data on the Nef mutants support the structural observations on CD4 binding and the involvement of the Nef N-terminal loop in the complex.
Nef binding induces a conformational change in the β2-μ2 half of AP2.
Overlaying the α and σ2 subunits of the “open” AP2 structure26 on the current structure reveals that the β2 and μ2 N-terminal domains (μ2-NTD) of AP2 in the current structure “move” out as one rigid body, causing a greater “opening” of the tetramer (Fig. 3a). In addition, a large portion of the β2 N-terminus, residues 1–88 encompassing the first four helices, is displaced from the rest of β2. If β2 had maintained its original fold, then its N-terminus would clash with the Nef core (Extended Data Fig. 2); the observed displacement is likely necessary to accommodate Nef-binding. While most of this displaced portion of β2 becomes disordered, the first helix preserves its helical structure and binds to the Nef core through hydrophobic interactions (Fig. 3bc).
Fig. 3. Nef binding induces conformational change in the β2 subunit of AP2.
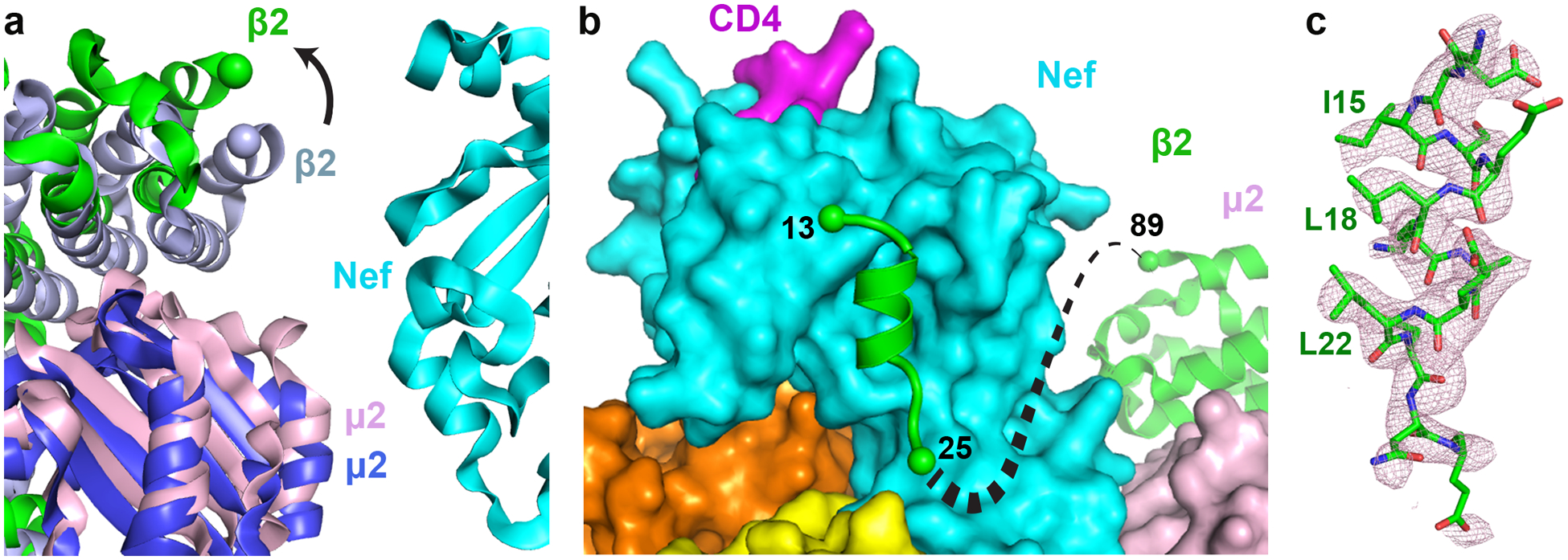
a, Current structure is overlaid, on the α-σ2 half, with the open AP2 (PDB ID: 2XA7). β2 (green) and μ2 (pink) subunits in the current structure move outward in comparison to β2 (light blue) and μ2 (blue) subunits of the unbound AP2. b, The first helix of β2 binds to the Nef core, while the next three helices become disordered. c, Density for the first helix of β2 (pink mesh, 2Fo-Fc map at 1.0σ with B factor sharpening of -50Å2) is shown. Residues important for association with Nef core are labeled.
The Nef-induced destabilization and structural changes of the β2-μ2 half of AP2 were further characterized by chemical cross-linking mass spectrometry (XL-MS) and integrative structure modeling (Extended Data Fig. 3). Here, disuccinimidyl sulfoxide (DSSO), a MS-cleavable, bifunctional amine-reactive small molecule, was used to cross-link proximal Lys residues or N-termini of the Nef-CD4CD fusion and AP2Δμ2-CTD complex. Cross-linked proteins separated by SDS-PAGE were trypsin digested and resulting peptides analyzed by specialized LC-MS3 experiments for identification of cross-linked residues (Extended Data Fig. 3a). Application of this pipeline to the complex identified intra- and inter-linked peptides corresponding to 90 unique cross-linked residues (Supplementary Table 1, Extended Data Fig. 3b). Lys residues from the displaced β2 N-terminal domain (1–88) are involved in a total of 24 cross-links (Supplementary Table 1, bold entries). Importantly, most of these cross-links are made by Lys residues flanking the first helix (i.e. Lys5, 11, 12, 26, and 27), consistent with this helix binding specifically in the complex. In contrast, Lys residues from the other dislocated β2 N-terminal helices (i.e. Lys29, 31, 35, 36, 45, 66, 67, 78) are rarely observed, consistent with a lack of fixed residence within the complex. The XL-MS data, which captures structural information from a conformationally heterogeneous population of protein complexes as they exist in solution, the crystal structure, and other structural information (Methods) were used for integrative modeling34 to produce a model ensemble which describes the complex in full (Extended Data Fig. 3c). Segments that are disordered in the crystal structure are modeled and represented as helices or flexible strings of beads (Extended Data Fig. 3c). The model ensemble agrees satisfactorily with the observed cross-links (89% of cross-links are satisfied, Extended Data Fig. 3d). Furthermore, it indicates large variability in the positions and orientations of the helices in the partially unfolded β2 segment (Extended Data Fig. 3e–g), consistent with the structural heterogeneity of this region indicated by the crystallographic data.
Nef uses a “molecular switch” to differentially downregulate MHC-I via AP1 or CD4 via AP2.
Binding of the first helix of β2 to the Nef core in the current structure closely resembles how the N-terminal amphipathic helix of Nef (deleted from the construct used herein) binds the same location when Nef hijacks AP1 for MHC-I downregulation (Fig. 4a)35. That interaction, mediated by Nef Trp13 and Met20, is critical for MHC-I downregulation but is dispensable for CD4 downregulation25,35,36. Our current structure indicates that intramolecular association of this Nef helix with the Nef core would force the Nef N-terminal loop to deviate from the conformation associated with CD4-binding (Extended Data Fig. 4). We suspect that this is an intricate plot of the virus: Nef binding forces the N-terminal helices of β2 to destabilize and unfold. By providing a binding-site for the first β2 helix on its core, Nef partially compensates for this destabilization. Moreover, the N-terminal β2 helix is now used to compete the Nef amphipathic helix off the core, freeing the N-terminal loop of Nef to adopt the conformation shown in Figure 2c for CD4 downregulation. A potential caveat to this hypothesis is that the L100A:I109A mutation of Nef, which should affect the binding of the β2 helix to the Nef-core (Fig. 4a), minimally impairs the downregulation of CD4 (Fig. 4b). However, since this mutation likely affects the binding of the β2 helix and the Nef N-terminal amphipathic helix similarly (Fig. 4a), it might have minimal influence on the competition between these two helices for the Nef core in vivo. In contrast, and consistent with the model above, the same mutation impairs the downregulation of class I MHC (Fig. 4c), presumably because it disrupts the required binding between the N-terminal amphipathic helix of Nef and the Nef core.
Fig. 4. The downregulation of MHC-I and CD4 are distinct both mechanistically and structurally, yet their cytoplasmic domains share a common binding site on Nef.
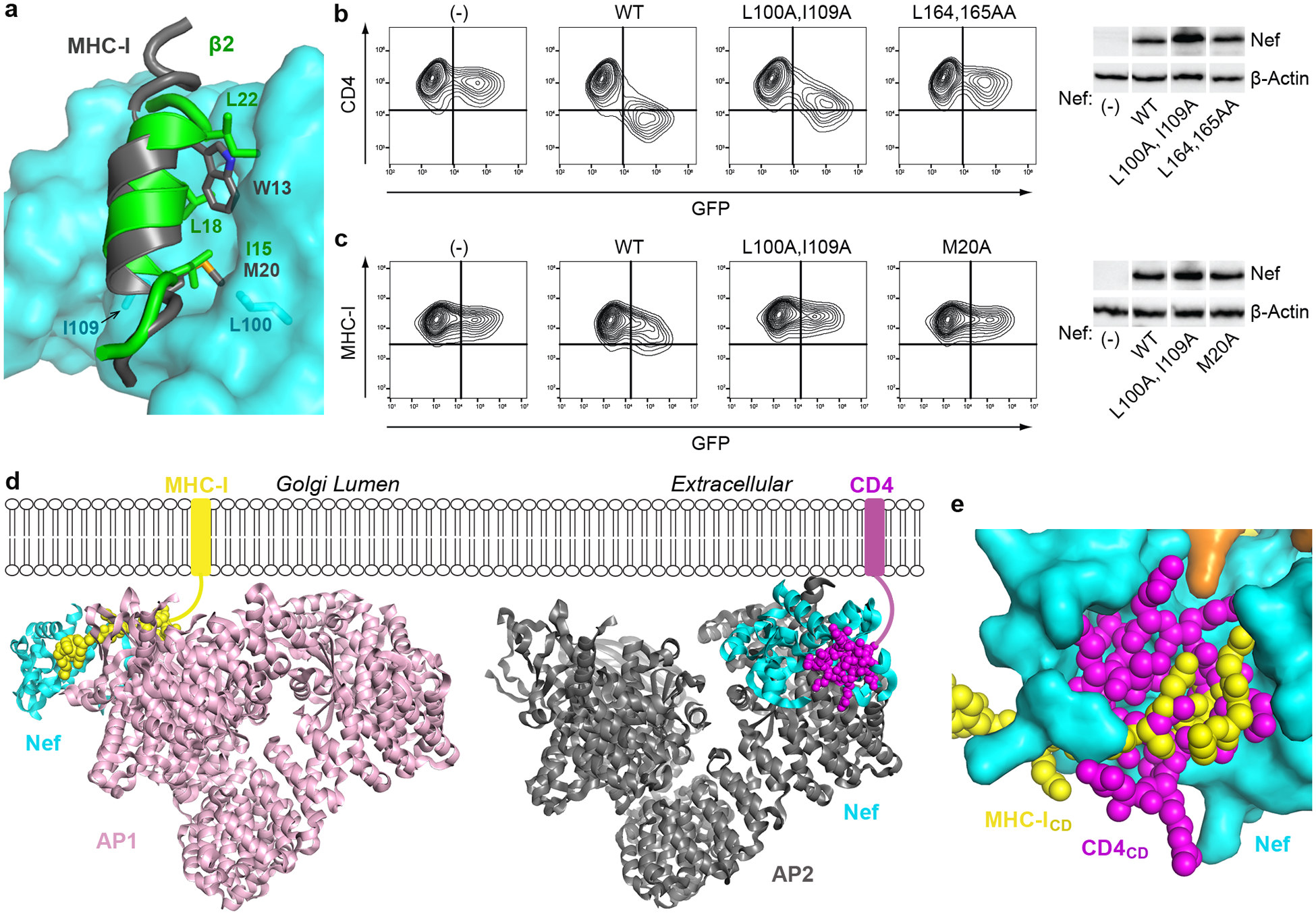
a, Nef/μ1/MHC-ICD (PDB 4EMZ) and Nef/AP2/CD4CD (current structure), are overlaid on Nef (cyan). The Nef N-terminal helix (gray) and the first helix of β2 (green, current structure) occupy the same site on the Nef core. Trp13 and Met20 residues in the Nef N-terminal helix and important for the intramolecular helix-core association are shown. Important residues on β2 (I15, L18, L22) are also shown. Nef L100 and I109 make direct contact with the bound helices. b, CD4 and c, MHC-I downregulation by Nef mutants was measured using cell surface-staining and flow cytometry; GFP is a transfection marker, GFP only control is indicated (-). Nef L164A:L165A and M20A mutants were used as negative controls for CD4 and MHC-I downregulation, respectively. CD4-positive HeLa cells were used for the CD4 assays, and HEK293 cells, which express HLA-A2, were used for the MHC-I assays. Each experiment was done at least twice (independent transfections). Cellular expression of Nef proteins was compared by western blot, the cellular control is β-actin. Uncropped blot images are shown in Supplementary Figure 1. d, Structural comparison of Nef/AP1/MHC-ICD (PDB 4EMZ; other AP1 subunits modeled based on overlay of the μ1-CTD) and Nef/AP2/CD4CD (current structure) shows that Nef hijacks AP1 (left) to downregulate MHC-I and AP2 (right) to downregulate CD4 in very different ways. e, Same overlay as in a. Binding of CD4CD (magenta) and MHC-ICD (yellow) involve the same pocket on Nef, yet, with the exception of Nef D123, the specific contact-residues are different.
Comparing the current structure with our earlier structure of Nef in complex with the μ1-CTD and the MHC-I cytoplasmic domain reveals versatility and specificity in how the structurally homologous AP1 and AP2 are selectively co-opted to downregulate MHC-I and CD4 (Fig. 4d). For MHC-I downregulation, Nef interacts solely with the μ1 subunit and exploits the conserved Tyr-based motif-binding site on AP135. For CD4 downregulation, Nef exploits the acidic dileucine-binding site on the α and σ2 subunits of AP2 and contacts all subunits except μ2. Our models show that by allowing the N-terminal Nef helix and the N-terminal β2 helix to bind the same pocket on the core, Nef creates a “molecular switch” that links its use of different AP complexes with the modulation of different targets: binding to AP2 frees the N-terminus of Nef from the core to recruit CD4, whereas binding to AP1 leaves the core free to bind the N-terminus of Nef and thereby facilitates the recruitment of MHC-I.
Despite these distinct modes of binding, the cytoplasmic domains of MHC-I and CD4 share much of the same binding “pocket” on Nef (Fig. 4e). As evident from the structural overlay, the two cytoplasmic domains partially overlap at this location. Notably, Asp123 is the only Nef residue in this pocket that is crucially involved in both binding mechanisms as well as required for the downregulation of both CD428,29 (Fig. 2b,e,f–h) and MHC-I28,29,35–37. Nonetheless, several residues at this site on Nef are highly conserved (Extended Data Fig. 5).
Discussion
Our new data enable us to postulate the sequence of events leading to the sequestration of CD4 by Nef. Nef first binds to AP2 using its C-terminal loop. This binding, involving the acidic dileucine motif-binding site of AP2, is extensive and robust (Fig. 1d). Unlike typical membrane cargos, which use only short cytoplasmic tails to bind AP2, Nef also contains a bulky core domain, which affixes to the α and σ2 half of AP2. Sterically, the bound Nef core is incompatible with an intact β2 subunit (Extended Data Fig. 2). Destabilization of β2 then occurs, causing the N-terminal helices to deviate from the bundled state (Fig. 3). The first helix of β2 becomes available to bind the Nef core, competing off the N-terminal helix of Nef and thereby freeing the rest of the Nef’s N-terminal loop (Fig. 4a). This loop then moves over, placing a short helical anchor at the interface between Nef and the α subunit and extending its arm to complete the CD4-binding site (Fig. 2).
Our results weigh against the notion that dimerization of Nef is required for CD4 downregulation. The dependence of CD4 downregulation on Asp123 has been previously attributed to its role in mediating Nef dimerization28. In addition, a quadruple Nef mutant, I109D:L112D:Y115D:F121D (ILYF), which cannot dimerize, is unable to downregulate CD432,38. However, as revealed here, a single Nef molecule is sufficient to recruit CD4CD into AP2. Moreover, the residues involved in dimerization are directly involved in critical elements of the model here. For example, Asp123 is in direct contact with CD4 (Fig. 2b). Three of the four residues involved in the quadruple mutant, Leu112, Tyr115, and Phe121, interact directly with either CD4 or Nef Trp57/Leu58 (Fig. 2bd). Finally, the ILYF-dependent dimerization of Nef, which occurs when Nef is in complex with the Src-family kinase Hck39, is incompatible with the conformation of the Nef N-terminal loop shown here as important for CD4 downregulation (Extended data Fig. 6).
Our results also weigh against the notion that the helix binding site on the Nef core is a promiscuous acceptor for the cytoplasmic domains of target proteins40. This hypothesis derived from the observation that this hydrophobic crevice is occupied by the acidic leucine-motif from an adjacent Nef molecule in a crystal structure of a simian immunodeficiency virus (SIV) Nef40. Portions of the cytoplasmic domains of CD4 and the CD3 ζ-chain could also be modeled into this site40. Our data and model reveal a very different scenario: this hydrophobic crevice on the Nef core binds two distinct helices: a helix from the β N-terminus when in complex with CD4 and AP2, or a helix from the Nef N-terminus when in complex with MHC-I and AP1 (Fig. 4a). As detailed above, this helix binding site on the Nef core is the basis for an elegant molecular switch mechanism that links the use of specific AP complexes with the modulation of different cellular targets.
A key feature of the β2 subunit revealed here is that it is structurally labile and, upon Nef-binding, can deviate from its original fold. Nef-induced destabilization of the β2 subunit has also been observed in a recently published structure describing how Nef from an SIV hijacks AP2 to recruit rhesus BST2 for surface downregulation41. There, the structural changes at the N-terminus of β2 occur in a completely different manner than here. Part of its first helix (14–17) loses its helical fold and instead refolds into a β sheet together with a preceding β2 segment (6–12) and a segment of Nef’s C-terminal loop. This β sheet then becomes part of the binding pocket for the recruitment of the rhesus BST2 tail. Whether such a conversion into a β sheet occurs when Nef, of either HIV or SIV, targets other cellular surface proteins via AP2-dependent endocytosis is unknown. Similarly, whether the conformational change in β2 observed in the present study (Fig. 3) is involved in other downregulation events mediated by Nef remains to be shown. Nonetheless, Nef clearly takes advantage of the destabilization that it causes in β2 to induce novel and distinct β2 conformations that facilitate recruitment of specific target proteins into clathrin-coated vesicles.
Antiretroviral drugs targeting Nef have the potential to unleash immunologic surveillance mechanisms. Our finding that the structural basis of the downregulation of MHC-I and CD4 involves a common and conserved binding site suggests that both activities could be inhibited by a single agent. Such a Nef inhibitor could facilitate the clearance of infected cells and contribute to HIV-cure-strategies42.
Methods
Materials
Gene of rat α adaptin is a kind gift from Dr. Juan Bonifacino (NIH). Genes of β2, μ2, and σ2 adaptins were amplified from a cDNA library of human HEK293T cells. For cross-linking mass spectrometry analysis, anhydrous dimethyl sulfoxide (DMSO), disuccinimidyl sulfoxide (DSSO), MS-grade trypsin, HPLC-grade water, formic acid, and acetonitrile were all purchased from Thermo Fisher Scientific. 4–20% TGX SDS-PAGE gels were purchased from Bio-Rad. MS-safe AcquaStain was purchased from Bulldog Bio.
Cloning, expression, and purification of proteins
The Nef-CD4CD fusion was constructed by fusing CD4 (394–419) to the C-terminus of HIV-1 Nef (26–206, NL4.3) via a flexible linker of 36 amino acids (GVDGSDEASELACPTPKEDG-LAQQQTQLNLRGSGSG). The encoding gene was cloned into a pMAT9 expression vector. The fusion protein was over-expressed in E. coli NiCo21(DE3) cells, carrying a N-terminal maltose binding protein (MBP) tag. Cells were induced with 0.1 mM isopropyl β-d-thiogalactopyranoside (IPTG) at OD600 of 0.8 and grown at 16°C overnight. Cells were lysed using sonication. Expressed protein was first purified using the MBP affinity column. For GST pull-down experiments, the MBP-tagged protein was further purified by a HiTrap Q anion exchange column, followed by a final Superdex 200 size exclusion column. For crystallization, the MBP fusion tag was cleaved off by the SARS-CoV Mpro protease43,44 after the affinity purification. The tagless protein was similarly purified using HiTrap Q anioin exchange column and the Superdex 200 size exclusion column. WT and mutant Nef proteins used for the FP assays were produced similarly.
For AP2Δμ2-CTD, genes for each of the four subunits were cloned into two duet vectors: a pETDuet vector with human β2(1–591) and human μ2(1–135) at each of the multiple cloning sites, respectively; a pCDFDuet vector with rat α(1–621) carrying a C-terminal GST tag and human σ2(1–142). The heterotetrameric AP2Δμ2-CTD core was expressed overnight at 22°C in the NiCo21(DE3) cells in Terrific broth after induction with IPTG. For binding assays, the GST-tagged the AP2Δμ2-CTD complex was purified by a sequence of Ni-NTA gravity column, GST affinity column, and Superdex 200 size exclusion column. For crystallization, the GST tag was cleaved off by the Tev protease after the GST affinity purification, followed by a final purification using the Superdex 200 size exclusion column.
In vitro GST pulldown assay
Purified proteins AP2Δμ2-CTD-GST (0.2 mg) and MBP-Nef-CD4CD (0.4 mg) were mixed in a final volume of 100 ul and incubated at 4°C for 30 minutes. The protein solution was then loaded onto a small gravity flow column containing 0.2 ml GST resin. Flow through was collected and the resin was extensively washed with 5 × 0.9 ml GST binding buffer (50 mM Tris, pH 8, 100 mM NaCl, 0.1 mM TCEP). The bound proteins were then eluted with 5 × 0.1 ml GST elution buffer containing 10 mM reduced glutathione. The eluted proteins were analyzed by SDS-PAGE.
Fluorescence polarization (FP) assay
Purified MBP-Nef and AP2Δμ2-CTD proteins were buffer exchanged, respectively, into the assay buffer (50 mM sodium phosphate, pH 6.5, 5 mM KCl, 2 mM Dithiothreitol). A stock protein solution was then prepared containing 80 μM MBP-Nef and 120 μM AP2Δμ2-CTD, which was subsequently used to create different dilutions. Assays were carried out in Corning 384-well black microplates (3820, Kennebunk, ME). In each well, 200 nM tetramethylrhodamine(TMR)-labeled CD4 peptide (CD4 408–417, synthesized by GenScript, Piscataway, NJ) was mixed with MBP-Nef and AP2Δμ2-CTD at varied concentrations in a total volume of 50 μl. Incubation was carried out for 1 hour at room temperature with minimal exposure to light. Fluorescence polarization was then measured using the EnVision plate reader (Perkin Elmer) with excitation at 535 nm and emission at 595 nm. FP measurement was repeated three times and plotted as a function of protein concentration in a logarithmic scale using Origin.
Crystallization and crystallographic data collection
Crystallization was carried out using the microbatch under-oil method. The purified AP2Δμ2-CTD core and the Nef-CD4CD fusion chimera were mixed at 1:5 molar ratio to a final concentration of 2.5 mg/ml (25 mM Tris, pH 8.0, 100 mM NaCl, 0.1 mM TCEP, 0.1 mM PMSF). Equal volumes of the protein solution and the precipitant solution (100 mM HEPES, pH 6.9, 200 mM KCl, 15% PEG4000, 6% 1,6-hexanediol) were mixed. The drop was sealed using a mixture of paraffin and silicon oil at a 2:1 ratio. Crystals appeared within 24 h at room temperature and grew to full size in about a week.
Crystals were cryo-protected using the precipitant solution containing 20% glycerol and then frozen in liquid nitrogen. Datasets were collected at NE-CAT (24-ID) at the Advanced Photon Source, Argonne National Laboratory, and FMX/AMX (17-ID) at the National Synchrotron Light Source II, Brookhaven National Laboratory. Diffraction data was processed using HKL200045. The crystals were in the P41 space group and diffracted to a highest resolution of 3.0 Å. The statistics are summarized in Table 1.
Structure determination and refinement
The structural solution was obtained by molecular replacement using PHASER46 in PHENIX47. Only one molecule exists in the asymmetric unit. The PDB of the open AP2 core (2XA7) was divided into two search models: the α and σ2 hemicomplex and the β2 and μ2-NTD hemicomplex. Together with the Nef structure (4EMZ), the three models were used sequentially to successfully obtain the solution. Iterative rounds of model building in COOT48 and refinement in Phenix49 were carried out. The final model has an Rwork/Rfree of 0.241/0.277. A Ramachandran plot showed that 96.4% of the residues are in the favored region, together with 3.5% in the allowed region and 0.1% as outliers. The refinement statistics are summarized in Table 1.
CD4 downregulation assays
HeLa cells expressing CD4 (TZM-bl, obtained from Dr. John Kappes via the NIH AIDS Reagent Program) were transfected using Lipofectamine2000 (Thermo Fisher Scientific) with pCG-GFP (a gift from Dr. Jacek Skowronski) and pCI-NL, a pCI-neo-based plasmid (Promega) expressing NefNL4–3 or the indicated Nef-mutants. 1.6 μg of total plasmid DNA was used in each transfection: 0.4 μg of pCG-GFP and 1.2 μg of pCI-NL or derivative-mutants. One of the replicate experiments was done using Fugene 6 (Promega) as the transfection reagent, 0.3 μg of pCG-GFP, and 0.9 μg of pCI-NL or derivative mutants. One day later, half the cells were stained for surface CD4 (anti-human CD4, BioLegend, conjugated directly to APC), fixed in formaldehyde, then analyzed by two-color flow cytometry using an Accuri 6 flow cytometer (Becton, Dickinson and Company). “Live cell” gates were set using untransfected cells; gates for GFP were set using cells transfected only with pCI-neo; and gates for CD4 were set using cells stained with an APC-conjugated antibody isotype control. Two-color contour plots were prepared using FlowJo™ Software (Becton, Dickinson & Company), Version 10.6.1. The other half of the cells were lysed in Laemmli buffer, and the proteins were resolved on 10% denaturing SDS-PAGE gels before transfer to polyvinylidene difluoride membranes. Nef was detected using a polyclonal antiserum raised to NL4–3 Nef in sheep (a gift from Dr. Celsa Spina, University of California San Diego). β-actin was detected using a murine monoclonal antibody (Sigma-Aldrich). Species-specific secondary antibodies conjugated to HRP were visualized using Western Clarity detection reagent (Bio-Rad). Chemiluminescence was recorded using a ChemiDoc Imager System (Bio-Rad). The cells were tested for mycoplasma using a PCR assay and were negative.
Class I MHC downregulation assays
HEK293 cells, which naturally express HLA-A2, were transfected using Lipofectamine2000 with pCG-GFP (0.4 μg) and pCI-NL, the indicated Nef-mutants, or the empty vector pCI-neo (1.2 μg). One day later, half the cells were stained for surface HLA-A2 (BioLegend, anti-HLA-A2 conjugated directly to APC), fixed in formaldehyde, then analyzed by two-color flow cytometry using an Accuri 6 flow cytometer. “Live cell” gates were set using untransfected cells; gates for GFP were set using cells transfected only with pCI-neo; and gates for HLA-A2 were set using cells stained with an APC-conjugated antibody isotype control. Two-color contour plots were prepared using FlowJo™ Software (Becton, Dickinson & Company), Version 10.6.1. The other half of the cells were processed for western blot as above. The cells were tested for mycoplasma using a PCR assay and were negative.
DSSO-based cross-linking mass spectrometry analysis (XL-MS)
Individual preparations of Nef-CD4CD-AP2Δμ2-CTD complex (2.8mg/mL and 0.7mg/mL at 1:5 Nef-CD4CD molar excess) were cross-linked using increasing molar ratios of DSSO (Thermo Fisher Scientific), for 5, 10, or 30 minutes at 4 or 37ᵒC. Cross-linked proteins were separated on 4–20% TGX gradient SDS-PAGE gels (Bio-Rad), stained with MS-safe AcquaStain (Bulldog Bio), and cross-linked product bands excised and submitted for in gel reduction, alkylation, and trypsin digestion. Extracted peptides were separated online by Thermo Easy nLC 1000 by reverse-phase HPLC (75 μm × 30 cm fused silica packed with 1.9-μm Reprosil-Pur C18 AQ resin (Dr. Maisch-GmbH) column), running a linear gradient of 5–30% B in 50min, 35–95% B in 5 min, and 95% B for 4 min at a flow rate of 300 nL/min (buffer A: 100% H2O/0.1% FA; buffer B: 100% ACN/0.1% FA). For each sample, XL-MS3 data was acquired on Thermo Orbitrap Elite using two similar data dependent acquisition experiments50 where a single acquisition cycle consisted of: 1) one full MS1 scan (350–1500 m/z, 120,0000 resolution, AGC target of 1×106); 2) top two data-dependent MS2 scans (15,000 resolution, AGC target of 5×104, normalized collision energy = 22%); and 3) top three (or four) MS3 scans (ion count target 104, normalized collision energy = 35%). Precursor ions (charge state ≥4+) were dynamically excluded for 20 seconds (tolerance of 10 ppm). Charge state and dynamic exclusion were applied to MS2 but turned off for MS3 acquisition.
Raw data was extracted to MGF format using MSConvert51, with MS3 data used for protein and peptide searches. Searches were performed by batch-tag feature of a locally installed version of Protein Prospector (v. 5. 19. 1, University of California San Francisco), with DSSO remnant mass modifications set as variable modifications (e.g. Alkene, Sulfenic-acid, and Thiol)50. Peptide reports were generated using the Search Compare feature of Protein Prospector, and dead-end, intra-linked, and inter-linked peptides identified by in house software program XL-Discoverer (part of new XLTools suite)52. Summarization and confidence assignment of inter-linked peptides was performed by in house scripts that reduce ambiguous assignments and distribute redundant counts.
Integrative structure modeling of the Nef-CD4CD-AP2Δμ2-CTD complex
We applied an integrative structural modeling approach34,53–55 to characterize the structure of the Nef-CD4CD-AP2Δμ2-CTD complex in solution, based on the crystal structure and the 90 DSSO cross-links. Integrative structure determination proceeded through the standard four stages53,54,56–59: 1) gathering data, 2) representing subunits and translating data into spatial restraints, 3) configurational sampling to produce an ensemble of structures that satisfies the restraints, and 4) analyzing and validating the ensemble structures and data. The integrative structure modeling protocol (i.e, stages 2, 3, and 4) was scripted using the Python Modeling Interface (PMI) package, a library for modeling macromolecular complexes based on our open-source Integrative Modeling Platform (IMP) package54, version 2.8 (https://integrativemodeling.org). Files containing the input data, scripts, and output results are available at https://github.com/integrativemodeling/Nef_CD4_AP2.
(1). Gathering data:
Modeling was based on the crystal structure, a comparative model of the β2 subunit 24–89 region built based on the AP2 structure26 using MODELLER60,61 and the 90 DSSO cross-links.
(2). Representing subunits and translating data into spatial restraints:
To maximize computational efficiency while avoiding using too coarse a representation, we represented the Nef-CD4CD-AP2Δμ2-CTD complex using a coarse-grained one residue per bead representation. The regions absent from the crystallographic structure and the comparative model were represented by a flexible string of beads corresponding to one residue each. To explore the positions and orientations of the Nef-CD4CD-AP2Δμ2-CTD components, we defined the following rigid bodies: α-σ2, β2(89–591)-μ2, Nef-CD4CD-β2(15–23), and the 4 helices in the partially unfolded β2 segment (29–43, 49–61, 64–78, and 81–86). With this representation in hand, we next translated the input information into spatial restraints as follows.
First, the 90 DSSO cross-links were used to construct a Bayesian term that restrained the distances spanned by the cross-linked residues62,63. The cross-link restraints were applied to the one residue-per-bead representation for the X-ray structure, comparative models, and flexible strings of beads. Second, to use the crystal structure of the hexamer as a template, we imposed “structural equivalence” distance restraints between pairs of residues closer than 8.0 Å across an interface between two rigid bodies, designed to restrain the model to resemble the template as much as possible. Third, excluded volume restraints were applied to all pairs of beads56,64. Fourth, we also applied the sequence connectivity restraint, using a harmonic upper bound on the distance between two consecutive beads in a subunit, with a threshold distance equal to four times the sum of the radii of the two connected beads56,64.
(3). Configurational sampling to produce an ensemble of structures that satisfies the restraints:
The initial positions and orientations of rigid bodies and positions of the beads in the flexible strings of beads were randomized. The structural models were generated using Replica Exchange Gibbs sampling, based on the Metropolis Monte Carlo (MC) algorithm63,65. Each MC step consisted of a series of random transformations (ie, rotations and translations) of the positions of the beads and rigid bodies. The sampling produced 4,000,000 models from 80 independent runs.
(4). Analyzing and validating the ensemble structures and data:
Model validation follows four major steps34,66:
(i) Selection of the models for validation: The ensemble of models for further analysis was objectively defined as follows. For each trajectory, the MC step at which all data likelihoods and priors are equilibrated (run equilibration step) was computed and all prior frames were discarded67. Sampling of the Nef-CD4CD-AP2Δμ2-CTD complex yielded 2,007,800 representative structures that sufficiently satisfied the input restraints.
(ii) Estimation of sampling precision: The precision at which sampling sampled the selected structures (sampling precision) was estimated66. The sampling precision must be comparable or higher than the precision of the structure ensemble consistent with the input data (model precision). As a proxy for testing the thoroughness of sampling, we performed four sampling convergence tests as described66. We performed three sampling convergence tests: 1) verify that the scores of refined structures do not continue to improve as more structures are computed. 2) Confirm that the selected structures in independent sets of sampling runs (Sample A and Sample B) satisfy the data equally well. The non-parametric Kolmogorov-Smirnov two-sample test (two sided) indicates that the difference between the two score distributions is insignificant (p-value (0.08)>0.05). In addition, the magnitude of the difference is small, as demonstrated by the Kolmogorov-Smirnov two-sample test statistic (D=0.02). 3) Cluster the structural models and determine the sampling precision at which the structural features can be interpreted. Three criteria were used for determining the sampling precision, evaluated as a function of the RMSD clustering threshold. First, the p-value is computed using the χ2-test (one-sided) for homogeneity of proportions. Second, an effect size for the χ2-test is quantified by the Cramer’s V value. Third, the population of structures in sufficiently large clusters (containing at least ten structures from each sample). Clustering is done at the RMSD threshold at which three conditions are satisfied (χ2-test p-value (1.0) > 0.05, Cramer’s V (0.0) < 0.10, and the population of clustered structures (0.96) > 0.80). The output of this protocol is a single distinct cluster containing the majority (96%) of the individual models. The sampling precision is defined as the average bead RMSD between the models within the cluster and its corresponding centroid in the finest clustering for which each sample contributes models proportionally to its size (considering both significance and magnitude of the difference) and for which a sufficient proportion of all models occur in sufficiently large clusters. The sampling precision for our integrative model is 13.5Å.
(iii) Estimation of model precision: The most explicit description of model uncertainty is provided by the set of all models that are sufficiently consistent with the input information (i.e. the ensemble). For example, if the models in the ensemble are clustered into a single cluster as in the case of the Nef-CD4CD-AP2Δμ2-CTD ensemble, the model precision is defined as the RMSD between models in the cluster. The precision for the Nef-CD4CD-AP2Δμ2-CTD model is 8.3 Å.
(iv) Quantification of the degree to which a model satisfies the information used to compute it: An accurate structure needs to satisfy the input information used to compute it. A DSSO cross-link restraint is satisfied by a cluster of models if the corresponding Cα–Cα distance in any of the models is less than 30 Å68. The ensemble satisfies 89% of the XLs used to compute it, including all of the Nef XLs (Extended Data Fig. 3). The unsatisfied crosslinks mostly span residues between CD4CD and the NTD’s of the α and β2 subunits (Supplementary Table 1). These violations can be rationalized as non-specific interactions between CD4CD and the AP2 complex when CD4CD is not bound to Nef in solution, false-positive crosslinks, sample heterogeneity, insufficient conformational sampling, and coarse-grained representation of the modeled components. The remaining restraints, including structural equivalence, excluded volume, and sequence connectivity restraints, are also satisfied within their uncertainties.
To compare the X-ray structure to the ensemble obtained using integrative modeling, we computed the distribution of the Cα root-mean-square deviation (RMSD) between the X-ray structure and each of the models in the ensemble. The mean Cα RMSD is 7.1 Å (4.3–12.2 95% CI).
To indicate the most flexible parts of the structure, we assessed the uncertainty of the position and orientation of each rigid body representing the hexamer in the model ensemble. To this end, all models were superimposed on each rigid body in turn, followed by computing the average RMSD for each of the other rigid bodies (Extended Data Fig. 3e). The model ensemble indicates large variability in the positions and orientations of the 4 helices in the partially unfolded β2 segment (Extended Data Fig. 3eg), consistent with the structural heterogeneity of this region indicated by the relative lack of electron density from crystallography.
Reporting Summary Statement:
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Code availability:
Files containing the input data, scripts, and output results for the integrative structure modeling of the Nef-CD4CD-AP2Δμ2-CTD complex are available at https://github.com/integrativemodeling/Nef_CD4_AP2.
Data Availability Statement:
The coordinates and structural factors for the crystal structure have been deposited at the Protein Data Bank (PDB) with the accession code 6URI. The proteomics XL-MS data has been deposited at the ProteomeXchange database69 with the accession code PXD019338. The integrative structural model has been deposited at PDB-Dev with the accession code PDBDEV_00000050. Uncropped gel images and source data for graphs are available with the paper online.
Extended Data
Extended Data Fig. 1. Electron density map for the N-terminal loop of Nef.
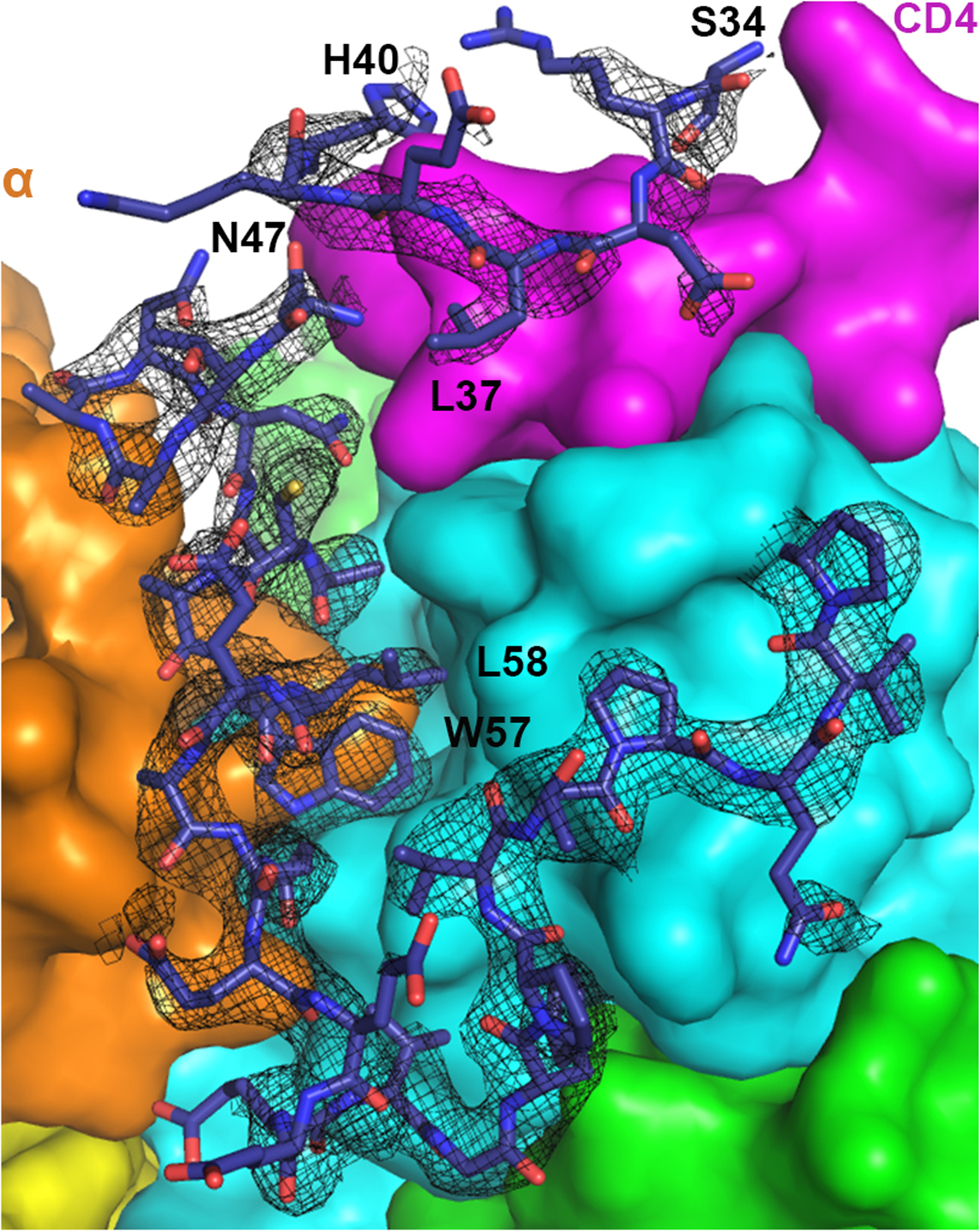
2Fo-Fc map (1σ level with B factor sharpened by -50Å2) for Nef residues 34–40 and 47–75 is shown as black mesh. Nef residues 41–46 could not be built due to the lack of density. Density for Nef 34–40 is less defined, although sidechain density for Leu37 is clear.
Extended Data Fig. 2. β2 subunit, if intact, would clash with the bound Nef.
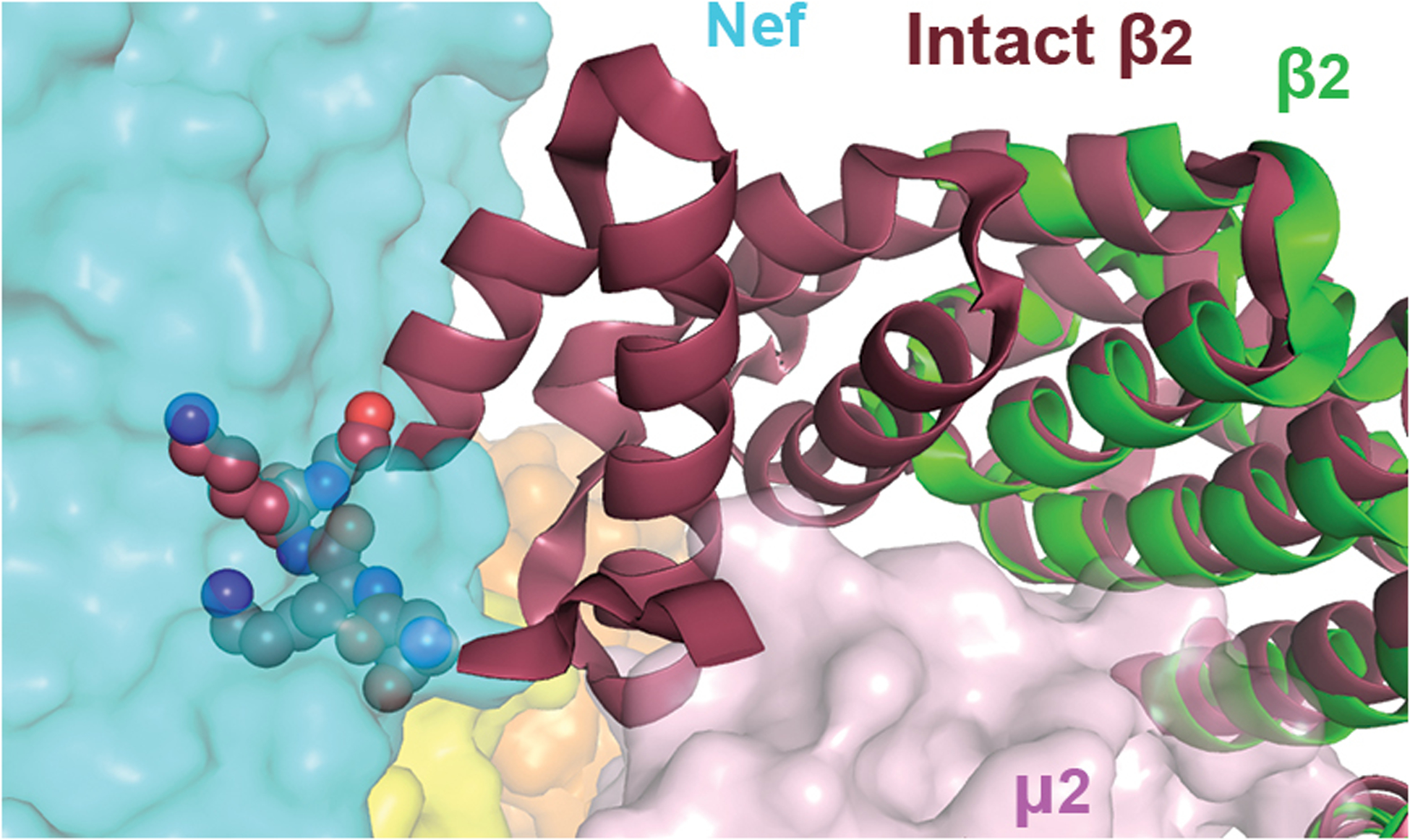
Overlay of the intact β2 subunit (dark red, PDB 2XA7) with β2 in the current structure (green) indicates that clashing would take place between Nef and N-terminus of the intact β2, specifically residues Asn10, Lys11, Lys12, and Gly13 (spheres).
Extended Data Fig. 3. Crosslinking mass spectrometry and integrative structure modeling of the Nef-CD4CD-AP2Δμ2-CTD complex.
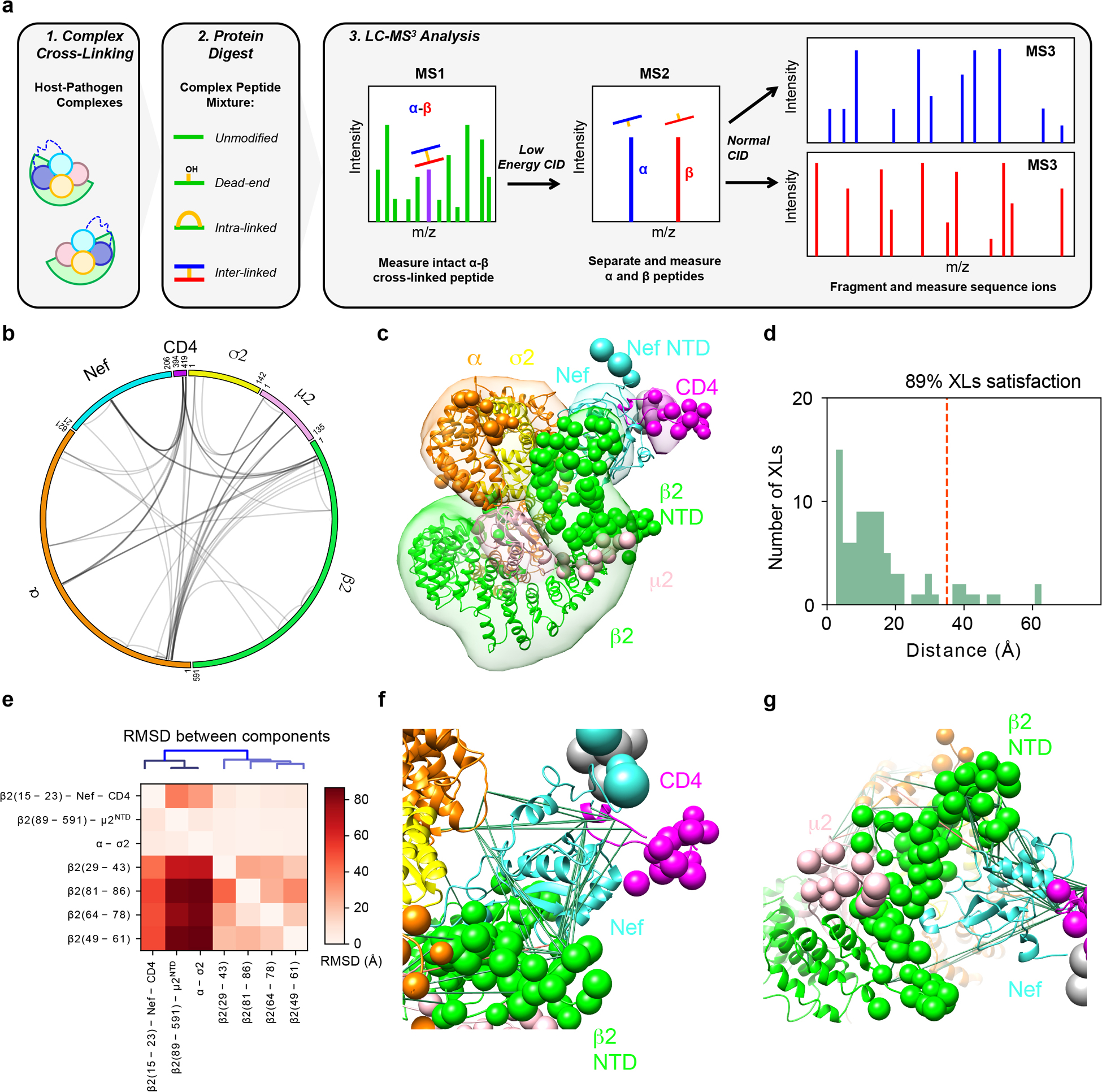
a, Overview of the DSSO XL-MS3 analysis method. b, CX-Circos linkage map of all Nef-CD4CD-AP2Δμ2-CTD interlinks. c, Integrative structure of the Nef-CD4CD-AP2Δμ2-CTD complex. The localization probability density of the ensemble of structures is shown with representative (centroid) structure from the ensemble embedded within it. Regions present in the crystal structure are shown as ribbons and segments not present in the crystal structure are shown as beads. d, Histogram showing the distribution of the cross-linked Cα–Cα distances in the integrative structure. The structural ensemble satisfies 89% of the XLs used to compute it. e, RMSD between rigid-bodies in the model ensemble. The vertical axis corresponds to the rigid body used as reference for superimposition and the horizontal axis are the rigid bodies for which the average RMSD was computed. f, Detail of crosslinks mapped to Nef. Satisfied and violated crosslinks shown in green and pink, respectively. g, Positioning of the unfolded β2 segment.
Extended Data Fig. 4. Binding of Nef N-terminal helix to the Nef core is incompatible with CD4 downregulation.
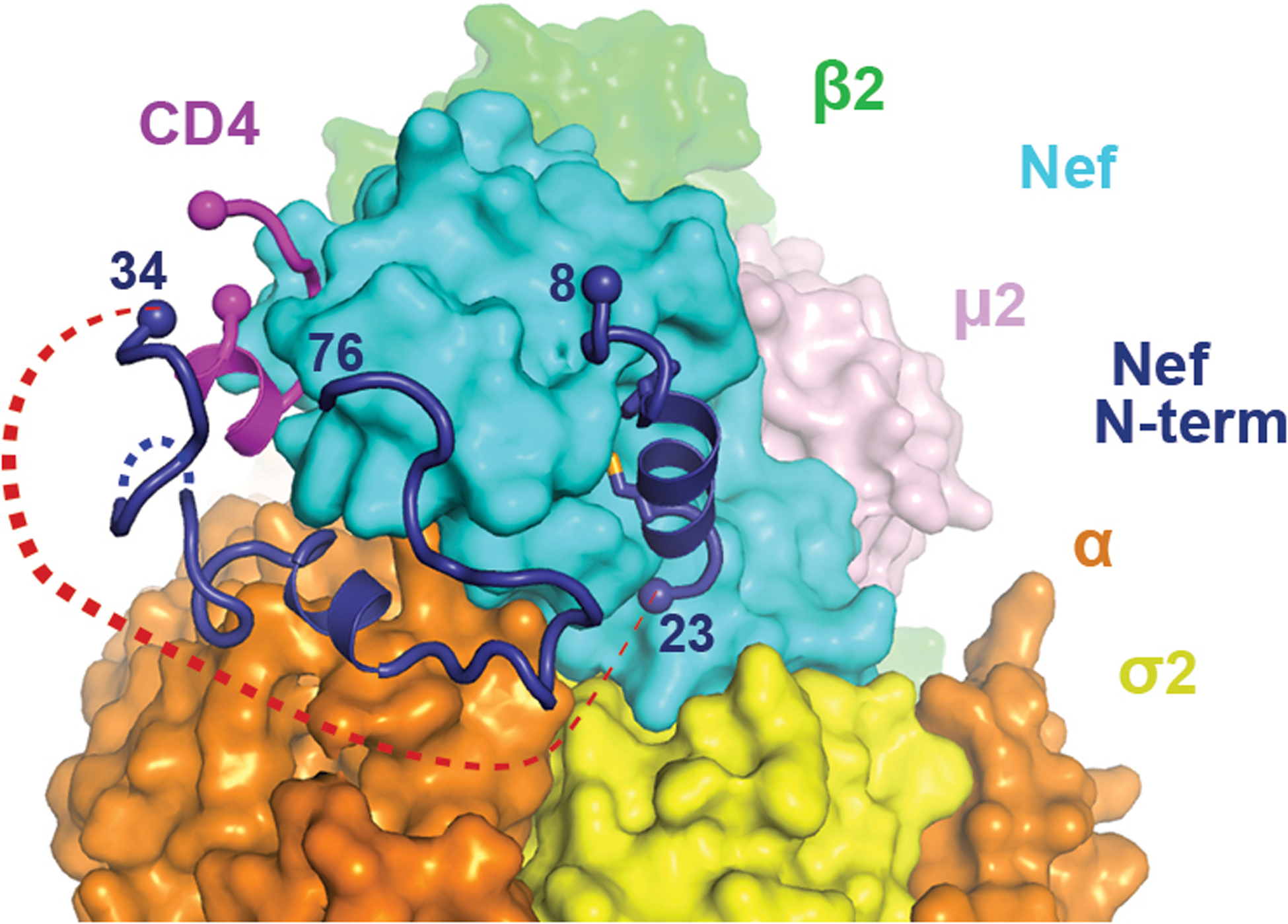
N-terminal helix of Nef (8–23) is modeled into the current structure. Red dotted line represents the would-be distance between residues 23 and 34, which cannot be covered by ten residues (Nef 24–33).
Extended Data Fig. 5. Nef residues at the CD4 -binding pocket are highly conserved.
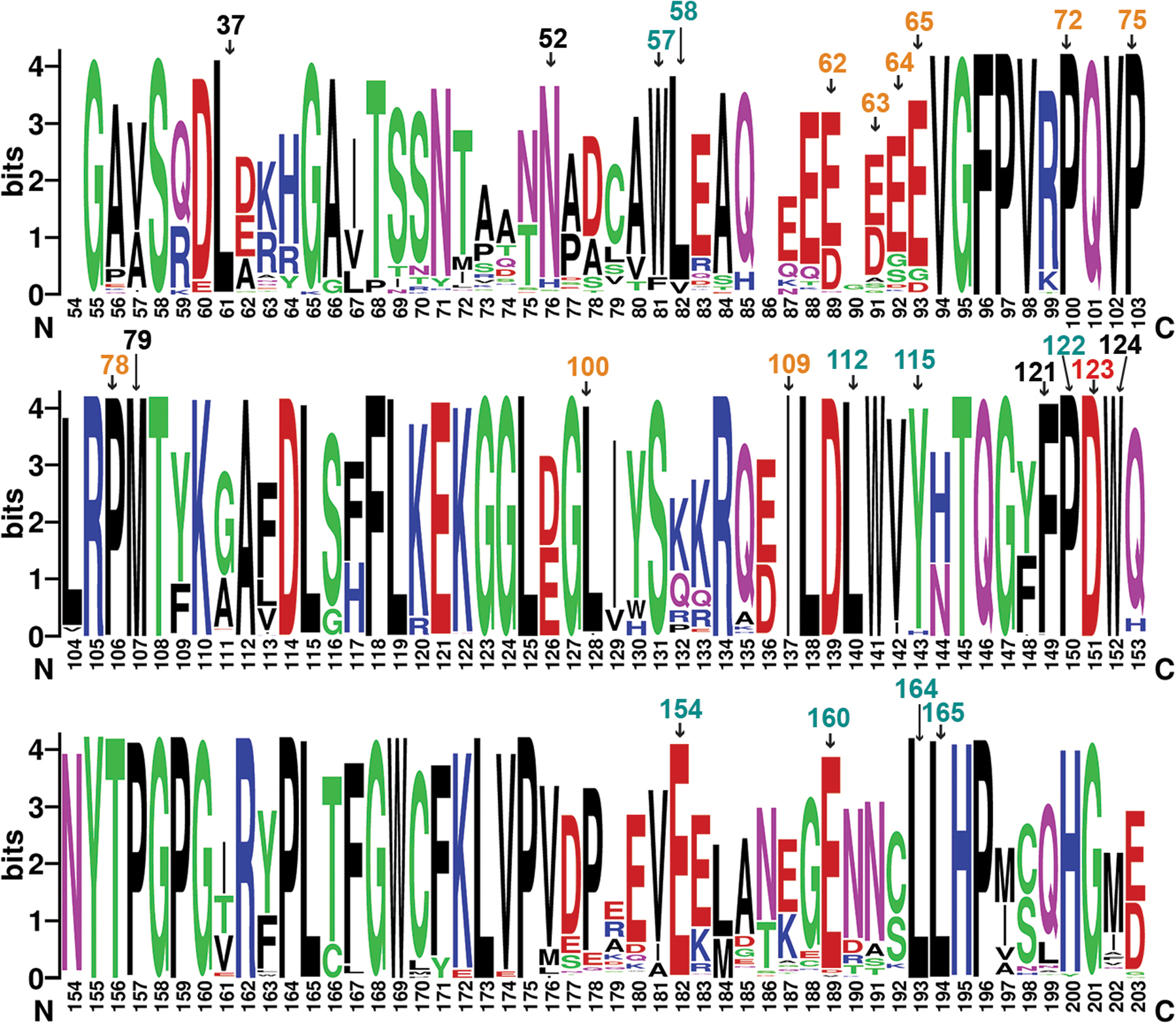
Nef sequences from HIV sequence compendium 2017 were analyzed though multiple sequence alignment (HIV sequence database, www.hiv.lanl.gov). Alignment was done in HXB2 convention (bottom) and important residues are additionally labeled using the NL4.3 convention on top. D123, shown in red text, is important for both CD4 and MHC-I downregulation. Other residues important for CD4 downregulation are in cyan and black texts. Black texts refer to residues, in addition to D123, that surround CD4. Other residues important for MHC-I downregulation are in orange. The logo representation, with the height of each letter proportional to the observed frequency of the corresponding amino acid residue, was generated by WebLogo70.
Extended Data Fig. 6. The unique conformation of Nef N-terminal loop observed in the current structure is incompatible with Nef dimerization.
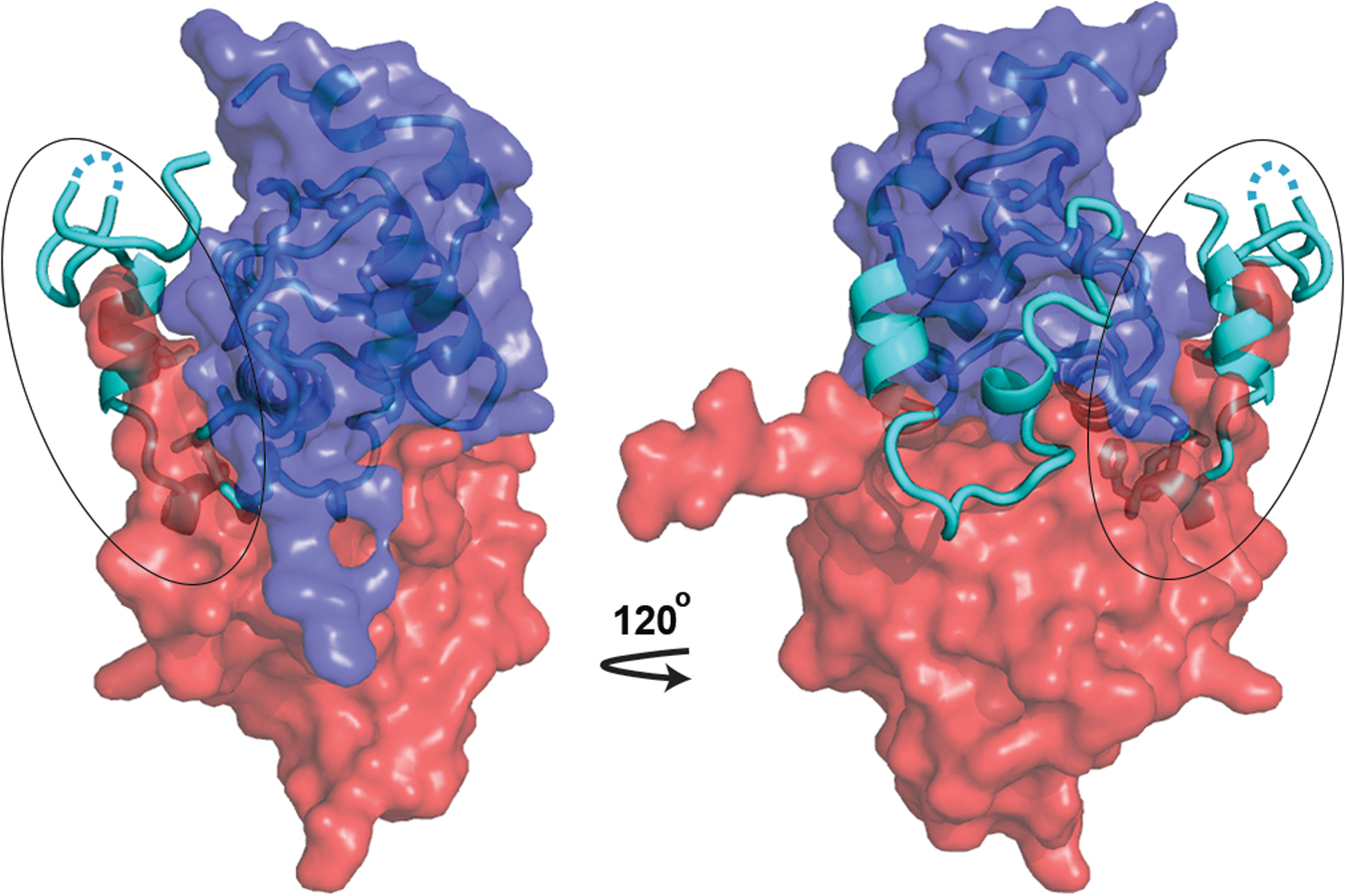
Nef in current conformation (cyan, cartoon) is overlaid with the SH2-SH3-dependent Nef dimer39 (dark blue and red envelopes, PDB 4U5W). While majority of Nef in the current structure overlays well with the Nef protomer shown as the dark blue envelope, the N-terminal region of Nef (circled) intrudes severely into the volume of the other Nef protomer (red envelope).
Supplementary Material
Acknowledgments:
We thank Yong Xiong (Yale University) for helpful discussions and valuable input. We thank the beamline staff at the Advanced Photon Source beamline 24-ID and the National Synchrotron Light Source beamline 17-ID. We thank Juan Bonifacino (NIH) for providing the gene of rat α adaptin. Funding: This work was supported by the University of Massachusetts Dartmouth startup fund (X.J.), US National Institutes of Health (NIH) grants AI102778 and AI129706 (J.G.). R.M.K., I.E., A.S., and N.K. were supported by NIH grant P50AI150476. R.M.K. was also supported by NIH fellowship F32AI127291. A.S. was also supported by NIH grants U19AI135990, R01GM083960, P41GM109824, and S10OD021596. N.K. was also supported by NIH grants P50GM082250 and U19AI135990.
Footnotes
Competing Interests Statement: The authors declare no competing interests.
References
- 1.Kirchhoff F, Schindler M, Specht A, Arhel N & Munch J Role of Nef in primate lentiviral immunopathogenesis. Cellular and Molecular Life Sciences 65, 2621–2636 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kestler HW 3rd et al. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 65, 651–62 (1991). [DOI] [PubMed] [Google Scholar]
- 3.Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL & Desrosiers RC Brief Report - Absence of Intact Nef Sequences in a Long-Term Survivor with Nonprogressive HIV-1 Infection. New England Journal of Medicine 332, 228–232 (1995). [DOI] [PubMed] [Google Scholar]
- 4.Deacon NJ et al. Genomic Structure of an Attenuated Quasi-Species of HIV-1 from a Blood-Transfusion Donor and Recipients. Science 270, 988–991 (1995). [DOI] [PubMed] [Google Scholar]
- 5.Pereira EA & daSilva LL HIV-1 Nef: Taking Control of Protein Trafficking. Traffic 17, 976–96 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Pawlak EN & Dikeakos JD HIV-1 Nef: a master manipulator of the membrane trafficking machinery mediating immune evasion. Biochim Biophys Acta 1850, 733–41 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Willey RL, Maldarelli F, Martin MA & Strebel K Human immunodeficiency virus type 1 Vpu protein regulates the formation of intracellular gp160-CD4 complexes. J Virol 66, 226–34 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross TM, Oran AE & Cullen BR Inhibition of HIV-1 progeny virion release by cell-surface CD4 is relieved by expression of the viral Nef protein. Curr Biol 9, 613–21 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Lama J, Mangasarian A & Trono D Cell-surface expression of CD4 reduces HIV-1 infectivity by blocking Env incorporation in a Nef- and Vpu-inhibitable manner. Curr Biol 9, 622–31 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Benson RE, Sanfridson A, Ottinger JS, Doyle C & Cullen BR Downregulation of cell-surface CD4 expression by simian immunodeficiency virus Nef prevents viral super infection. J Exp Med 177, 1561–6 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veillette M et al. Interaction with cellular CD4 exposes HIV-1 envelope epitopes targeted by antibody-dependent cell-mediated cytotoxicity. J Virol 88, 2633–44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pham TN, Lukhele S, Hajjar F, Routy JP & Cohen EA HIV Nef and Vpu protect HIV-infected CD4+ T cells from antibody-mediated cell lysis through down-modulation of CD4 and BST2. Retrovirology 11, 15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding S et al. CD4 Incorporation into HIV-1 Viral Particles Exposes Envelope Epitopes Recognized by CD4-Induced Antibodies. J Virol 93(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schubert U et al. CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway. J Virol 72, 2280–8 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.daSilva LLP et al. Human Immunodeficiency Virus Type 1 Nef Protein Targets CD4 to the Multivesicular Body Pathway. Journal of Virology 83, 6578–6590 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aiken C, Konner J, Landau NR, Lenburg ME & Trono D Nef Induces CD4 Endocytosis - Requirement for a Critical Dileucine Motif in the Membrane-Proximal Cd4 Cytoplasmic Domain. Cell 76, 853–864 (1994). [DOI] [PubMed] [Google Scholar]
- 17.Garcia JV & Miller AD Serine Phosphorylation-Independent Down-Regulation of Cell-Surface CD4 by Nef. Nature 350, 508–511 (1991). [DOI] [PubMed] [Google Scholar]
- 18.Guy B et al. HIV F/3’ orf encodes a phosphorylated GTP-binding protein resembling an oncogene product. Nature 330, 266–9 (1987). [DOI] [PubMed] [Google Scholar]
- 19.Chaudhuri R, Lindwasser OW, Smith WJ, Hurley JH & Bonifacino JS Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J Virol 81, 3877–90 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenberg ME et al. Co-localization of HIV-1 Nef with the AP-2 adaptor protein complex correlates with Nef-induced CD4 down-regulation. EMBO J 16, 6964–76 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Traub LM & Bonifacino JS Cargo recognition in clathrin-mediated endocytosis. Cold Spring Harb Perspect Biol 5, a016790 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren X, Park SY, Bonifacino JS & Hurley JH How HIV-1 Nef hijacks the AP-2 clathrin adaptor to downregulate CD4. Elife 3, e01754 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Preusser A, Briese L, Baur AS & Willbold D Direct in vitro binding of full-length human immunodeficiency virus type 1 Nef protein to CD4 cytoplasmic domain. Journal of Virology 75, 3960–3964 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salghetti S, Mariani R & Skowronski J Human immunodeficiency virus type 1 Nef and p56lck protein-tyrosine kinase interact with a common element in CD4 cytoplasmic tail. Proc Natl Acad Sci U S A 92, 349–53 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mangasarian A, Piguet V, Wang JK, Chen YL & Trono D Nef-induced CD4 and major histocompatibility complex class I (MHC-I) down-regulation are governed by distinct determinants: N-terminal alpha helix and proline repeat of Nef selectively regulate MHC-I trafficking. Journal of Virology 73, 1964–1973 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson LP et al. A Large-Scale Conformational Change Couples Membrane Recruitment to Cargo Binding in the AP2 Clathrin Adaptor Complex. Cell 141, 1220–U213 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hua J & Cullen BR Human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus Nef use distinct but overlapping target sites for downregulation of cell surface CD4. J Virol 71, 6742–8 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu LX et al. Mutation of a conserved residue (D123) required for oligomerization of human immunodeficiency virus type 1 Nef protein abolishes interaction with human thioesterase and results in impairment of Nef biological functions. Journal of Virology 74, 5310–5319 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hua J, Blair W, Truant R & Cullen BR Identification of regions in HIV-1 Nef required for efficient downregulation of cell surface CD4. Virology 231, 231–8 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Baugh LL, Garcia JV & Foster JL Functional characterization of the human immunodeficiency virus type 1 Nef acidic domain. Journal of Virology 82, 9657–9667 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aiken C, Krause L, Chen YL & Trono D Mutational analysis of HIV-1 Nef: identification of two mutants that are temperature-sensitive for CD4 downregulation. Virology 217, 293–300 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Poe JA & Smithgall TE HIV-1 Nef dimerization is required for Nef-mediated receptor downregulation and viral replication. J Mol Biol 394, 329–42 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaudhuri R, Mattera R, Lindwasser OW, Robinson MS & Bonifacino JS A Basic Patch on alpha-Adaptin Is Required for Binding of Human Immunodeficiency Virus Type 1 Nef and Cooperative Assembly of a CD4-Nef-AP-2 Complex. Journal of Virology 83, 2518–2530 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rout MP & Sali A Principles for Integrative Structural Biology Studies. Cell 177, 1384–1403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia X et al. Structural basis of evasion of cellular adaptive immunity by HIV-1 Nef. Nat Struct Mol Biol 19, 701–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams M, Roeth JF, Kasper MR, Filzen TM & Collins KL Human immunodeficiency virus type 1 Nef domains required for disruption of major histocompatibility complex class I trafficking are also necessary for coprecipitation of Nef with HLA-A2. Journal of Virology 79, 632–636 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wonderlich ER, Williams M & Collins KL The tyrosine binding pocket in the adaptor protein 1 (AP-1) mu 1 subunit is necessary for nef to recruit AP-1 to the major histocompatibility complex class I cytoplasmic tail. Journal of Biological Chemistry 283, 3011–3022 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Shu ST, Emert-Sedlak LA & Smithgall TE Cell-based Fluorescence Complementation Reveals a Role for HIV-1 Nef Protein Dimerization in AP-2 Adaptor Recruitment and CD4 Co-receptor Down-regulation. J Biol Chem 292, 2670–2678 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alvarado JJ, Tarafdar S, Yeh JI & Smithgall TE Interaction with the Src homology (SH3-SH2) region of the Src-family kinase Hck structures the HIV-1 Nef dimer for kinase activation and effector recruitment. J Biol Chem 289, 28539–53 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manrique S et al. Endocytic sorting motif interactions involved in Nef-mediated downmodulation of CD4 and CD3. Nat Commun 8, 442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buffalo CZ et al. Structural Basis for Tetherin Antagonism as a Barrier to Zoonotic Lentiviral Transmission. Cell Host Microbe 26, 359–368 e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deeks SG HIV: Shock and kill. Nature 487, 439–40 (2012). [DOI] [PubMed] [Google Scholar]
Method-only References
- 43.Xue XY et al. Production of authentic SARS-CoV M-pro with enhanced activity: Application as a novel tag-cleavage endopeptidase for protein overproduction. Journal of Molecular Biology 366, 965–975 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue XY et al. Structures of two coronavirus main proteases: Implications for substrate binding and antiviral drug design. Journal of Virology 82, 2515–2527 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otwinowski Z & Minor W Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–26 (1997). [DOI] [PubMed] [Google Scholar]
- 46.McCoy AJ et al. Phaser crystallographic software. J Appl Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D-Biological Crystallography 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Afonine PV et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68, 352–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaake RM et al. A new in vivo cross-linking mass spectrometry platform to define protein-protein interactions in living cells. Mol Cell Proteomics 13, 3533–43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kessner D, Chambers M, Burke R, Agus D & Mallick P ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gutierrez CB et al. Developing an Acidic Residue Reactive and Sulfoxide-Containing MS-Cleavable Homobifunctional Cross-Linker for Probing Protein-Protein Interactions. Anal Chem 88, 8315–22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim SJ et al. Integrative structure and functional anatomy of a nuclear pore complex. Nature 555, 475–482 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russel D et al. Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol 10, e1001244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ward AB, Sali A & Wilson IA Biochemistry. Integrative structural biology. Science 339, 913–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alber F et al. Determining the architectures of macromolecular assemblies. Nature 450, 683–94 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Lasker K et al. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci U S A 109, 1380–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sali A et al. Outcome of the First wwPDB Hybrid/Integrative Methods Task Force Workshop. Structure 23, 1156–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schneidman-Duhovny D, Pellarin R & Sali A Uncertainty in integrative structural modeling. Curr Opin Struct Biol 28, 96–104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sali A & Blundell TL Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234, 779–815 (1993). [DOI] [PubMed] [Google Scholar]
- 61.Webb B & Sali A Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Protein Sci 86, 2 9 1–2 9 37 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Erzberger JP et al. Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex. Cell 159, 1227–1228 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi Y et al. Structural characterization by cross-linking reveals the detailed architecture of a coatomer-related heptameric module from the nuclear pore complex. Mol Cell Proteomics 13, 2927–43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shen MY & Sali A Statistical potential for assessment and prediction of protein structures. Protein Sci 15, 2507–24 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swendsen RH & Wang JS Replica Monte Carlo simulation of spin glasses. Phys Rev Lett 57, 2607–2609 (1986). [DOI] [PubMed] [Google Scholar]
- 66.Viswanath S, Chemmama IE, Cimermancic P & Sali A Assessing Exhaustiveness of Stochastic Sampling for Integrative Modeling of Macromolecular Structures. Biophys J 113, 2344–2353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chodera JD A Simple Method for Automated Equilibration Detection in Molecular Simulations. J Chem Theory Comput 12, 1799–805 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Merkley ED et al. Distance restraints from crosslinking mass spectrometry: mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci 23, 747–59 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perez-Riverol Y et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Crooks GE, Hon G, Chandonia JM & Brenner SE WebLogo: a sequence logo generator. Genome Res 14, 1188–90 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structural factors for the crystal structure have been deposited at the Protein Data Bank (PDB) with the accession code 6URI. The proteomics XL-MS data has been deposited at the ProteomeXchange database69 with the accession code PXD019338. The integrative structural model has been deposited at PDB-Dev with the accession code PDBDEV_00000050. Uncropped gel images and source data for graphs are available with the paper online.


