SUMMARY:
DDX3X is frequently mutated in the WNT and SHH subtypes of medulloblastoma - the commonest malignant childhood brain tumor. But whether DDX3X functions as a medulloblastoma oncogene or tumor suppressor gene is not known. Here, we show that Ddx3x regulates hindbrain patterning and development by controlling Hox gene expression and cell stress signaling. In mice predisposed to Wnt or Shh-medulloblastoma, Ddx3x sensed oncogenic stress and suppressed tumor formation. WNT and SHH-medulloblastomas normally arise only in the lower and upper rhombic lips, respectively. Deletion of Ddx3x removed this lineage restriction, enabling both medulloblastoma subtypes to arise in either germinal zone. Thus, DDX3X is a medulloblastoma tumor suppressor that regulates hindbrain development and restricts the competence of cell lineages to form medulloblastoma subtypes.
Keywords: medulloblastoma, tumor suppressor gene, DDX3X, inflammasome, stress granule
Graphical Abstract

eTOC
DDX3X is frequently mutated in medulloblastoma - a common childhood hindbrain tumor. Patmore et al., show that Ddx3x regulates hindbrain development and suppresses medulloblastoma formation by controlling Hox gene expression, cell stress signaling and restricting the number of cell lineages that can form tumors.
INTRODUCTION
Genome-wide studies of cancer have transformed our understanding of the disease. Taxonomies based solely on tumor location and histology have evolved into more precise classifications that include details of the cancer transcriptome, methylome and mutational landscape (Doherty et al., 2019; Roychowdhury and Chinnaiyan, 2014). The hope is that this molecular revolution will usher in a new era of precision cancer medicine that ultimately improves patient outcome. But early successes in leukemia have proved hard to replicate in other malignancies (Druker et al., 2001; Letai, 2017). A deeper understanding of the functional consequences of genomic-aberrations is likely to be required if we are to fully realize the promise of precision medicine for all patients with cancer.
The challenge of translating genomic understanding into patient cures is well illustrated by medulloblastoma - the commonest malignant childhood brain tumor. Once considered a single disease, medulloblastoma is now known to comprise at least four subtypes, each with distinct origins, genomic drivers and clinical outcomes (Northcott et al., 2019): SHH-medulloblastomas arise from cerebellar granule neuron precursor cells (NPCs), contain activating mutations in the SHH pathway, and are fatal in ~25% of cases; Group 3-medulloblastomas likely arise from Nestin+ cerebellar NPCs, amplify MYC in 17% of cases, and kill around half of all patients; while Group 4 medulloblastomas probably arise within the cerebellar unipolar brush cell lineage, contain more complex genetic drivers and are fatal in ~25% of cases (Hovestadt et al., 2019; Jones et al., 2012; Kawauchi et al., 2012; Northcott et al., 2012b; Pugh et al., 2012; Robinson et al., 2012; Vladoiu et al., 2019; Yang et al., 2008). In stark contrast, WNT-medulloblastomas arise from mossy fiber NPCs in the lower rhombic lip, are curable even when metastatic, and frequently contain activating mutations in CTNNB1 (Gajjar et al., 2006; Gibson et al., 2010; Jessa et al., 2019; Northcott et al., 2012a; Northcott et al., 2011). While this knowledge has enabled more accurate prediction of patient prognosis and use of conventional treatment, it is yet to yield novel curative therapies. Achieving this will require an understanding of how genomic patterns are laid down and function in medulloblastoma, and in particular why specific driver mutations are exquisitely restricted to each tumor subtype.
The discovery that DDX3X - an RNA-binding protein of the DEAD-box family - is frequently mutated in WNT- (36%) and SHH- (21%) but not Group 3- or 4-medulloblastomas, provides an inroad to understand the functional basis of medulloblastoma subtypes (Jones et al., 2012; Northcott et al., 2012b; Pugh et al., 2012; Robinson et al., 2012). Therefore, we studied the role of DDX3X in hindbrain development and medulloblastoma tumorigenesis.
RESULTS
Ddx3x regulates hindbrain development
As a first step to understand the role of Ddx3x in health and disease, we generated a conditional knock-out allele (Ddx3xFlx) by inserting LoxP sites into introns 6 and 14 (Samir et al., 2019). Since Ddx3x is located on chromosome X, Cre-recombination of Ddx3xFlx led to the complete loss of Ddx3x protein from female Ddx3xFlx/Flx and male Ddx3xFlx/Y NPCs, and a ~50% reduction of expression in female Ddx3xFlx/+ NPCs (Figure 1A).
Figure 1. Ddx3x regulates hindbrain development.

A. Western blot of Ddx3x and actin loading control expression in Cre-recombined mouse NPCs of the indicated genotype. B. Phenotype-free survival of Blbp-Cre+/−;Ddx3xFlx/Flx (n=55), Blbp-Cre+/−;Ddx3xFlx/+ (n=29) and Blbp-Cre+/−;Ddx3xFlx/Y mice (n=31). C. Heatmaps reporting the log-fold difference in the indicated histological parameters quantified by automated photo-micromorphometry. Analyses were performed on ≥10 mice per time point for each indicated variable and genotype. D. Photomicrographs of morphology (hematoxylin and eosin stain) and Ddx3x RNA expression (in situ hybridization) in developing hindbrains of the indicated genotype (unless indicated scale bars=100μm). E. Microanatomy of the cerebellar cortex cells and associated markers used in F-H are: EGL=external germinal layer; IGL=internal granule cell layer; BG=Bergman glia; PC=Purkinje cell; SC=stellate cell. F. Photomicrographs of morphology (hematoxylin and eosin stain) of the P13 cerebellar cortex in the indicated mouse genotype (scale bars=100μm). G, H. In situ hybridization (Atoh1) and immunofluorescence of the indicated cell markers in P13 mouse cerebellar cortex of the indicated genotype. I. Ddx3y fragments per kilobase of transcript per million mapped reads in mouse hindbrain (n=3 to 5 per genotype, sex and age; *=p<0.05, Mann-Whitney). See also Figure S1.
Mice harboring Ddx3xFlx were bred with those carrying the Blbp-Cre allele that drives Cre-recombination from embryonic day (E) 9.5 in NPCs throughout the neuroaxis, including lower and upper rhombic lip NPCs that are competent to form Wnt- and Shh-medulloblastomas, respectively (Gibson et al., 2010). Thirty-eight percent (n=21/55) of female Blbp-Cre+/−;Ddx3xFlx/Flx mice displayed severe ataxia and seizures within 34 days of life (Figure 1B). Growth of all brain regions, especially the cerebellum, was significantly impaired in these animals relative to Blbp-Cre+/−;Ddx3x+/+ control mice (≥10 mice per time point; Figure 1C) and was associated with induction of apoptosis (cleaved Caspase 3), cell cycle arrest (p21), and/or loss of total cell number (Figure 1C; Figure S1A). Blbp-Cre+/−;Ddx3xFlx/Flx mice had morphologically normal cerebella at E16.5 but subsequently displayed progressive abnormalities of both upper and lower rhombic lip-derived cell lineages, including: loss of normal cerebellar foliation and lamination (Figure 1D–F); failure of formation of major brainstem nuclei (Figure S1B); collections of abnormal ‘rests’ of proliferating Atoh1+/Pax6+/Ki67+ cerebellar granule NPCs intermingled within sheets of Synaptophysin+ neurons (Figure 1G and H); disorganization of the normal Gfap+ Bergman glia radial scaffold (Figure 1H); and haphazard positioning of Calbindin+ Purkinje cells (Figure 1H). Brain growth and cell organization were also disrupted in Blbp-Cre+/−;Ddx3xFlx/Flx mice that survived into adulthood, albeit to a lesser extent than those displaying neurological impairment (data not shown). In contrast, neither Blbp-Cre+/−;Ddx3xFlx/+ female (n=29; median follow-up=101 days) nor Blbp-Cre+/−;Ddx3xFlx/Y male mice (n=31; median follow-up=182 days) displayed neurological, brain size or cell organization abnormalities; although the total number of cells throughout the brains of these mice was reduced at P30, suggesting that loss of Ddx3x is not compensated for completely in these animals (Figure 1B–H).
Thus, Ddx3x specifies and/or maintains hindbrain lineages, including those that generate Wnt and Shhmedulloblastoma. Although Ddx3x expression is ablated completely in NPCs of Blbp-Cre+/−;Ddx3xFlx/Y male mice (Figure 1A), these mice developed relatively normally; therefore, the paralog Ddx3y likely compensates for Ddx3x during development in a manner similar to that of other paralog pairs (Welstead et al., 2012). In keeping with this notion, Ddx3y was upregulated in Blbp-Cre+/−;Ddx3xFlx/Y hindbrains relative to male Blbp-Cre+/−;Ddx3x+/+ controls (P13 and P30 P<0.05; Figure 1I).
Ddx3x regulates hindbrain patterning and stress response
Hindbrain development involves a highly-orchestrated program of regional gene expression. Central to this process are the Hox genes that govern the formation of distinct rhombomeres (Kiecker and Lumsden, 2005). Since Ddx3x regulates global patterns of gene expression, including the Hox genes, we looked to see if Hox expression is perturbed in the hindbrain following Ddx3x deletion (Duijkers et al., 2019; Herdy et al., 2018; Valentin-Vega et al., 2016).
Global patterns of gene expression among developing Blbp-Cre+/−;Ddx3x+/+, Blbp-Cre+/−;Ddx3xFlx/+, Blbp-Cre+/−;Ddx3xFlx/Y and Blbp-Cre+/−;Ddx3xFlx/Flx hindbrains were dictated predominantly by developmental stage rather than genotype (≥5 hindbrains per timepoint and genotype; Figure 2A). However at E16.5, 0.08% (27/32,075) and 0.12% (38/32,075) of all RNA transcripts were upregulated and downregulated respectively in Blbp-Cre+/−;Ddx3xFlx/Flx hindbrains relative to age matched Blbp-Cre+/−;Ddx3x+/+ controls (Figure 2B). Expression of these genes was not affected in Blbp-Cre+/−;Ddx3xFlx/+ or Blbp-Cre+/−;Ddx3xFlx/Y hindbrains. Remarkably, 58% (22/38) of downregulated genes are Homeobox genes (FDR=3.2e−23), including: Gbx1 that specifies the midbrain-hindbrain boundary; Pax2 and Gbx2 (primordial cerebellum); Hoxa2 (rhombomere 2); Hoxb2 (rhombomere 3); Hoxa3 and Hoxb3 (rhombomeres 5 and 6); Hoxa4 and Hoxb4 (rhombomere 7); and Phox2b, Tbx20, and Nkx6–1 (regulators of developmental gene transcription; Figure 2B). Pathway analysis of all dysregulated genes using g:GOST within the g:Profiler interface (Raudvere et al., 2019) and Cytoscape algorithms (Shannon et al., 2003) confirmed enrichment for regulators of neuronal development (n=42 related pathways; FDR=4.7e−2 to 3.2e−23), RNA processing and transcription (n=11 related pathways; FDR=1.1e−3 to 6.9e−9), cranial nerve development (n=3 related pathways; FDR=2.8e−5 to 2.0e−7) and cell motility (n=1 related pathway; FDR=3.8e−2; Figure 2B). By P5, Blbp-Cre+/−;Ddx3xFlx/Flx hindbrains also expressed lower levels of neural fate commitment genes including Ascl1, Kirrel2, Lhx5, Neurog1, Neurog2 and Pax2 that mark cerebellar ventricular zone NPCs and GABAergic lineages (Figure 2B): Lhx5 is a known Hox target gene (Makki and Capecchi, 2011). These data suggest strongly that Ddx3x regulates early hindbrain development by directly, or indirectly, regulating Hox gene expression and that Ddx3y compensates for Ddx3x loss during development, at least in part, by maintaining Hox expression.
Figure 2. Ddx3x regulates tissue patterning and stress response in the hindbrain.

A. Principal components analysis of RNAseq profiles generated from hindbrains isolated from the indicated mice (≥5 hindbrains per timepoint and genotype). B. Centre, heat map of the log-fold difference of expression of the indicated genes in hindbrains of mice with the indicated genotypes relative to Blbp-Cre+/−;Ddx3x+/+ controls (≥5 hindbrains per timepoint and genotype at the indicated developmental time points). Left, Cytoscape™ plots identifying gene sets with related functions that are significantly up- or down-regulated in Blbp-Cre+/−;Ddx3xFlx/Flx hindbrains relative to Blbp-Cre+/−;Ddx3x+/+ controls at the indicated developmental stage. Right, in situ hybridization of the indicated genes in Blbp-Cre+/−;Ddx3xFlx/Flx and Blbp-Cre+/−;Ddx3x+/+ hindbrains. Cartoons indicate the location of the higher power images (scale bars=100μm). See also Figure S2.
We showed previously that Ddx3x shuttles between stress granules and NLRP3-inflammasomes to coordinate live-or-die decisions in response to cell stress (Samir et al., 2019). Incorporation of Ddx3x into stress granules promotes a cell survival signal, while its inclusion in inflammasomes drives pyroptosis - a form of programmed cell death important in the innate immune response and neurodegeneration (McKenzie et al., 2019a; Voet et al., 2019). In keeping with this function, P13 Blbp-Cre+/−;Ddx3xFlx/Flx hindbrains dysregulated inflammasome (FDR=1.1e−6), cell stress (n=2 related pathways; FDR=1.1e−4 to 1.0e−6), immune (n=16 related pathways; FDR=0.05 to 6.0e−5) and apoptotic response regulators (n=5 related pathways; FDR=0.01 to 5.6e−5; Figure 2B). Histological review demonstrated that this response was triggered in neuronal cells rather than representing a significant infiltration of immune cells or expansion of resident microglia (Figure S2). Thus, aberrant development of the postnatal Blbp-Cre+/−;Ddx3xFlx/Flx hindbrain might be exacerbated by deregulation of stress granule-inflammasome signaling in neuronal cells in a manner similar to that reported in neurodegenerative conditions (McKenzie et al., 2019a; Voet et al., 2019). No such response was observed in Blbp-Cre+/−;Ddx3xFlx/Y mice despite the fact that Ddx3y does not compensate for the stress signaling functions of Ddx3x (Samir et al., 2019). Therefore, we propose that abnormal development of the embryonic Blbp-Cre+/−;Ddx3xFlx/Flx hindbrain, rather than deletion of Ddx3x per se, triggers stress signals to which these mice are unable to respond appropriately.
Ddx3x is a medulloblastoma tumor suppressor gene
We next sought to understand the role of DDX3X in medulloblastoma. WNT and SHH pathway mutations are found almost exclusively in WNT and SHH-medulloblastomas respectively, while mutations in DDX3X occur frequently in both of these tumor subtypes (Northcott et al., 2019). The molecular basis of this subtype-specific distribution of mutations and whether DDX3X is a medulloblastoma oncogene or tumor suppressor gene is not known. Therefore, we recorded patterns of tumor formation in mice harboring the Ddx3xFlx allele that were predisposed to develop either Wnt- (Blbp-Cre+/−;Ctnnb1Flx/+;Tp53Flx/+, hereon mWnt) or Shh-medulloblastoma (Blbp-Cre+/−;Ptch1Flx/+, hereon mShh).
As expected (Gibson et al., 2010), 7% (n=4/55, median surveillance=145 days) of mWnt;Ddx3x+/+ mice developed medulloblastomas embedded in the brainstem with no evidence of transformation in the cerebellum (Figure 3A, C and H). mWnt;Ddx3xFlx/+ mice also developed tumors exclusively from the dorsal brainstem with a similar incidence and latency (Figure 3A and H). In stark contrast, 27% (n=13/47) of mWnt;Ddx3xFlx/Y and 7% (n=3/44) of mWnt;Ddx3xFlx/Flx mice developed tumors that were confined solely to the cerebellum with no evidence of tumorigenesis in the brainstem - presumably because of the longer latency of brainstem relative to cerebella tumorigenesis. Tumors in these mice ranged from a diffuse expansion of the external granule cell layer to discrete tumor masses (Figure 3D and E). Both comprised highly proliferative, Pax6+/Ki67+/Atoh1+/Lef1+ small round blue cells, compatible with mutant Ctnnb1-driven transformation of granule NPCs. Notably, nuclear G3bp1 expression that has been associated with cell stress (Tourrière et al., 2001) was particularly upregulated in cerebellar mWnt;Ddx3xFlx/Y medulloblastomas (Figure 3D).
Figure 3. Ddx3x is a medulloblastoma tumor suppressor gene.
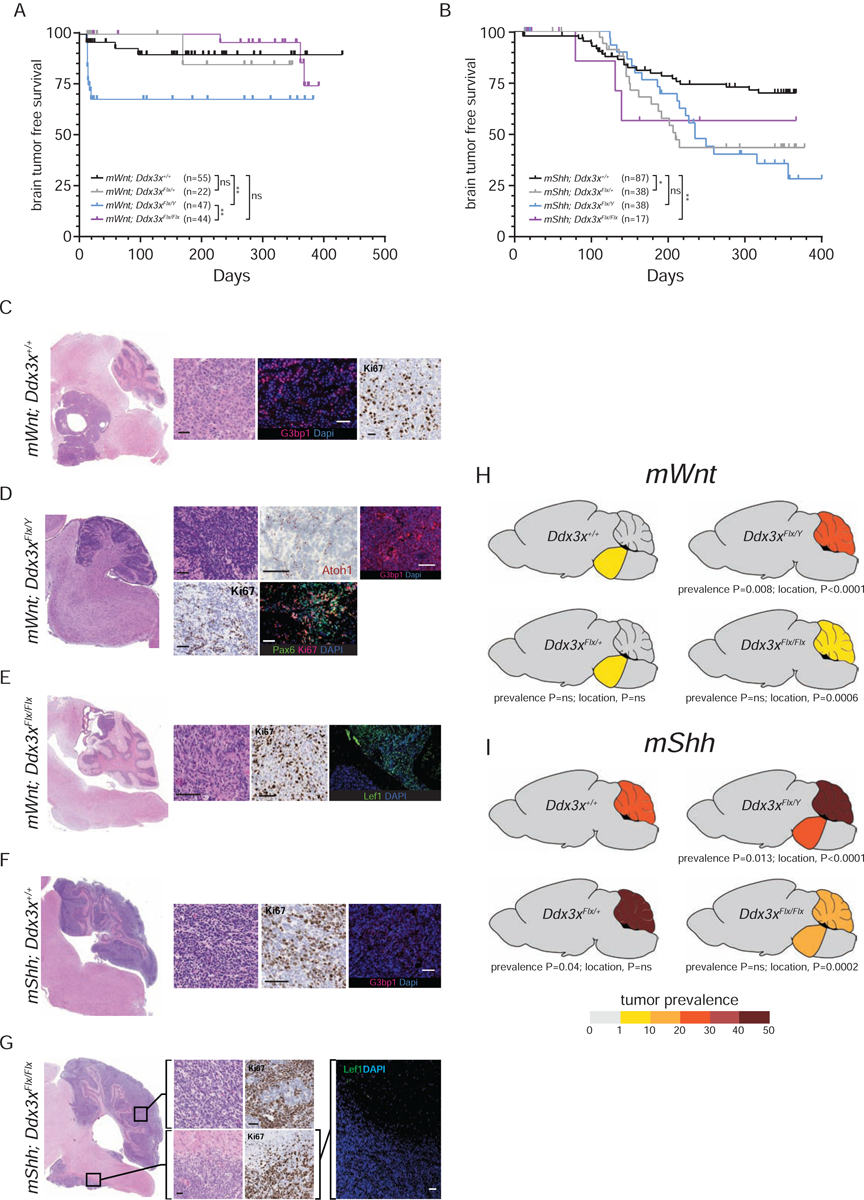
Tumor-free survival curves of mice predisposed to develop either Wnt- (A) or Shh-medulloblastoma (B) harboring the indicated Ddx3x alleles. Numbers of mice ‘n’ are shown for each cohort. ns=not significant, *=p<0.05, **=p<0.005, Log-Rank statistic. Photomicrographs of low and high-power images of tumors arising in mice predisposed to develop Wnt- (C-E) or Shh-medulloblastomas (F,G) harboring the indicated Ddx3x alleles. High power images reveal tumor morphology (hematoloxylin and eosin), proliferation (Ki67 immunohistochemical stain), or immunofluorescence of the indicated markers. T=tumor, (scale bars=100μm). Cartoons depicting the incidence and distribution (brainstem or cerebellum) of tumors arising in mice predisposed to develop either Wnt- (H) or Shh-medulloblastoma (I) harboring the indicated Ddx3x alleles. Mouse numbers same as in (A and B). See also Figure S3.
Remarkably, Ddx3x also suppressed both the prevalence and distribution of mShh tumors. Twenty-six percent (n=23/87) of mShh;Ddx3x+/+ mice developed tumors that were confined solely to the cerebellum (Figure 3B, F and I). Medulloblastomas also formed exclusively in the cerebella of mShh;Ddx3xFlx/+ mice, although with a significantly greater prevalence and reduced latency (45%, n=17/38; prevalence, P=0.04; survival Log Rank, P<0.05; Figures 3B and I). In contrast, of the 50% (n=19/38) of mShh;Ddx3xFlx/Y mice that developed tumors, more than half arose in the brainstem (Figure 3B and I). In two mice these tumors were confined solely to the brainstem, while eight arose simultaneously in the brainstem and cerebellum. Of the 18% (n=3/17) of mShh;Ddx3xFlx/Flx mice that developed tumors, two occurred simultaneously in the cerebellum and brainstem (Figure B, G and I). mShh;Ddx3x+/+ and mShh;Ddx3xFlx/Y cerebella tumors preserved the locus and expression of the wild-type Ptch1 allele, in keeping with reports the Ptch1 serve as a haploinsufficient suppressor of medulloblastoma (Wetmore et al., 2000) (Figure S3). All mShh tumors, including those in the brainstem, were Lef1− distinguishing them from tumors driven by activation of Ctnnb1 (Figure 3G). Similar to the development of mWnt tumors, the prevalence and location of mShh tumors were impacted most significantly in male Ddx3xFlx/Y mice (Figure 3B and I).
WNT-medulloblastomas have not been reported to arise in the cerebellum nor SHH-medulloblastomas in the brainstem, suggesting that Ddx3x restricts the competence of cell lineages to form these tumor subtypes. However, it is also possible that these tumors had spread directly, or metastatically, from their more recognized origins. Therefore, we sought additional evidence that mWnt;Ddx3xFlx/Y cerebellar tumors are distinct from those in the brainstem of mWnt;Ddx3x+/+ mice (we were unable to isolate adequate material for further analysis from mShh;Ddx3xFlx/Y or mShh;Ddx3xFlx/Flx brainstem, or mWnt;Ddx3xFlx/Flx cerebellar tumors). RNAseq analysis identified three very discrete groups of tumors: (i) all mShh cerebellar tumors regardless of Ddx3x genotype; (ii) mWnt;Ddx3x+/+ brainstem tumors; and (iii) mWnt;Ddx3xFlx/Y cerebellar tumors (Figure 4 A and B). As expected, mWnt;Ddx3x+/+ brainstem and all mShh cerebellar tumors recapitulated the gene expression signatures of their corresponding human medulloblastoma subtypes (Gibson et. al., 2010; Figure 4B). In keeping with their high-expression of Lef1, mWnt;Ddx3xFlx/Y tumors also displayed a human WNT-medulloblastoma signature (Figure 4B); however, in contrast to mWnt;Ddx3x+/+ brainstem tumors, mWnt;Ddx3xFlx/Y tumors potently upregulated cell cycle (n=34 related pathways; FDR=0.02 to 2.8e−44), immune (n=23 related pathways; FDR=0.04 to 9.0e−07), cell stress (n=4 related pathways; FDR=0.02 to 2.0e−19) and death response genes (n=18 related pathways; FDR=0.02 to 3.2e−7; Figure 4B and C). This pattern of gene expression was similar to that observed in the developing Blbp-Cre+/−;Ddx3xFlx/Flx hindbrain (Figure 2B). The transcriptome of mWnt;Ddx3xFlx/Y (and all mShh) cerebellar tumors was also distinguished from that of mWnt;Ddx3x+/+ brainstem tumors by upregulation of genes expressed by cerebellar granule NPCs and DNA repair pathways (n=11 related pathways; FDR=7.2e−4 to 1.6e−29) that are recognized features of human and mouse cerebella-derived medulloblastomas (Ratnaparkhe et al., 2018).
Figure 4. Ddx3x restricts cell lineage origins of Wnt-medulloblastoma.

A. Principal component analysis of RNAseq profiles generated from 30 mouse tumors (shown in B) arising in mice predisposed to develop Wnt- or Shh-medulloblastomas harboring the indicated Ddx3x alleles. B. Unsupervised hierarchical clustering of RNAseq profiles of 30 tumors shown in (A). Heatmap shows the log2 normalized expression values of the indicated genes. Gene set classification is shown left, including signature genes of WNT and SHH medulloblastoma. C. Cytoscape™ plot identifying gene sets with related functions that are significantly up- or down-regulated in mWnt;Ddx3xFlx/Y relative to mWnt;Ddx3x+/+ tumors shown in A and B. D. Tumor growth recorded as bioluminescence in mice implanted with cells isolated from cerebella of the indicated genotype (number of mice implanted with each cell type is shown ‘n’; ***=p<0.0005, Mann-Whitney). E. Photomicrographs of tumor (T) arising in the cerebellum of a recipient mice implanted with cerebellar cells isolated from mWnt;Ddx3xFlx/Y mice. Images reveal tumor morphology (hematoloxylin and eosin), proliferation (Ki67 immunohistochemical stain), or Lef1 immunofluorescence (scale bars=100μm). See also Figure S4.
To provide further evidence that the genomes of mWnt;Ddx3xFlx/Y cerebellar and mWnt;Ddx3x+/+ brainstem tumors are distinct, we performed shallow whole exome sequencing of DNA isolated from these tumors. Both analyzed mWnt;Ddx3xFlx/Y cerebella tumors, but neither the mWnt;Ddx3x+/+ brainstem nor two mShh;Ddx3x+/+ cerebella tumors, deleted a common region of 7qB3 (Figure S4). Conversely, gain of 6qA1–6qB3 and 18qA1–18qE4 and deletion of 17qE2–17qter were observed only in the mWnt;Ddx3x+/+ brainstem tumor but no mWnt or mShh cerebella tumors.
Finally, to determine directly if mWnt;Ddx3xFlx/Y cerebella can generate tumors, we isolated single cell suspensions from these tissues at P13 - prior to discernable tumor formation (n=12, dissected carefully away from the brainstem) - as well as P13 mWnt;Ddx3x+/+ (n=5), mWnt;Ddx3xFlx/Flx (n=3), Blbp+/−;Ddx3x+/+ (n=3), Blbp+/−;Ddx3xFlx/+ (n=3), Blbp+/−;Ddx3xFlx/Y (n=3), and Blbp+/−;Ddx3xFlx/Flx (n=3) cerebella. Cell suspensions were transduced with lentiviral-luciferase and 5×105 cells injected orthotopically into the cerebella of separate P28 immunocompromised mice as described previously (Mohankumar et al., 2015). Ki67+/Lef1+ tumors formed rapidly in all mice engrafted with mWnt;Ddx3xFlx/Y cells (Figures 4D and E). No other engrafted cell types formed tumors, including those from mWnt;Ddx3x+/+ cerebella that never formed tumors de novo or mWnt;Ddx3xFlx/Flx cerebella that formed tumors with very low penetrance (Figure 3H; median follow up 90 days).
Thus, Ddx3x suppresses the penetrance and restricts lineage origins of mWnt and likely mShhmedulloblastomas, providing a potential explanation why the gene is mutated in both of these tumor subtypes in humans. Since tumor formation was most penetrant in mWnt;Ddx3xFlx/Y and mShh;Ddx3xFlx/Y mice, then in contrast to its development functions, Ddx3y does not compensate for the tumor suppressor function of Ddx3x.
Ddx3x suppresses oncogenic Wnt-signaling in the cerebellum
To understand how Ddx3x might restrict cell lineage susceptibility to medulloblastoma tumorigenesis, we observed the impact of oncogenic signaling in developing mWnt (E16.5-P13) and mShh (E16.5-P30) Ddx3x+/+, Ddx3xFlx/+, Ddx3xFlx/Y and Ddx3xFlx/Flx cerebella relative to age-matched Blbp-Cre;Ddx3x+/+ controls prior to any evidence of tumor formation (≥5 cerebellar/genotype/time point). Global patterns of gene expression in the cerebella of these animals was dictated largely by developmental stage rather than genotype. However, all mWnt cerebella, including mWnt;Ddx3x+/+ and mWnt;Ddx3xFlx/+ that do not form tumors, displayed aberrant and robust Wnt-signaling that increased with development (Figure 5A,B). Thus, the absence of tumorigenesis in mWnt;Ddx3x+/+ and mWnt;Ddx3xFlx/+ cerebella does not result from an inability of these tissues to support aberrant Wnt signaling. Conversely, no mWnt cerebella, including mWnt;Ddx3xFlx/Y and mWnt;Ddx3xFlx/Flx that form tumors, displayed aberrant Shh-signaling. Thus, the formation of tumors in mWnt;Ddx3xFlx/Y and mWnt;Ddx3xFlx/Flx cerebella is unlikely to result from oncogenic Shh-signaling (Figures 5A and B).
Figure 5. Ddx3x suppresses oncogenic Wnt-signaling in the developing cerebellum.

Heatmaps reporting the log-fold difference (LFD) in expression of the indicated genes (A) or False Discovery Rate of the indicated pathway up- or down-regulation (B) in premalignant cerebellar isolated from mice predisposed to develop either Wnt- (left) or Shh-medulloblastoma (right) harboring the indicated Ddx3x alleles at the indicated developmental time points relative to Blbp-Cre+/−;Ddx3x+/+ controls (≥5 cerebellar/genotype/time point). Fold change in Ki67 (C), p21 (D) and cleaved Caspase 3 (E) immunohistochemistry in the mice shown in (A and B) relative to Blbp-Cre+/−;Ddx3x+/+ controls (n≥10). The number ‘n’ of brains analyzed in each assay is shown above each bar. *=p<0.05; **=p<0.005; ***=p<0.0005, Mann-Whitney. F. Exemplary immunohistochemical stains in P5 cerebella of the indicated mice; scale bar = 50μm.
Rather, compatible with an oncogenic-stress response, Wnt-signaling triggered a profound and progressive activation of cell stress (P13, FDR=8.7e−4 to 2.3e−10) cell death (P13, FDR=0.04 to 2.7e−9) and upregulation of p21 and cleaved Caspase 3 in mWnt;Ddx3x+/+ and mWnt; Ddx3xFlx/+ cerebella: DNA repair (P5, FDR=1.8e−12), cell cycle progression (P5, FDR=1.2e−7 to 1.5e−57) and cell proliferation (Ki67) were down-regulated concurrently in these mice (Figure 5A–F). In stark contrast, deletion of Ddx3x blunted mWnt-induced cell stress (P13, FDR=0.04 to 1.1e−6) and cell death (P13, FDR=0.01 to 0.003) markedly in mWnt;Ddx3xFlx/Y cerebella and neither pathway was induced in mWnt;Ddx3xFlx/Flx cerebella (Figure 5B). Furthermore, neither mWnt;Ddx3xFlx/Y nor mWnt;Ddx3xFlx/Flx cerebella downregulated DNA repair or cell cycle progression pathways and by P13 displayed normal levels Ki67, p21 and cleaved Caspase 3 (Figure 5C–E). Together, these data support the notion that aberrant Wnt signaling in the developing cerebellum provokes a potent oncogenic-stress response mediated by Ddx3x to suppress medulloblastoma tumorigenesis. Deletion of Ddx3x partially relieves this oncogenic-stress, enabling Wnt-driven malignant transformation. We are investigating if a reciprocal mechanism suppresses oncogenic Shh-signaling in the lower rhombic lip.
In contrast, aberrant Shh-signaling activated cell cycle progression, DNA repair, cell proliferation (Ki67) and a robust SHH-medulloblastoma signature (FDR=1.7e−5 to 3.5e−5) with minimal impact on p21 and cleaved Caspase 3 expression in all mShh cerebella, regardless of Ddx3x genotype, in keeping with the uniform transformation of these tissues (Figures 3I and 5A–F). These data support the notion that Shh signaling - the native mitogen for granule NPCs - is relatively well tolerated by the granule lineage, leading to unopposed transformation independent of Ddx3x status. Upregulation of cell cycle progression, DNA repair genes and cell proliferation (Ki67) was much less robust in mShh;Ddx3xFlx/Flx cerebella compatible with the relatively low level of tumorigenesis in these tissues (Figures 3I and 5B,C).
Stress signaling in DDX3X mutant human WNT-medulloblastomas
As a first step to determine if deletion of Ddx3x in mouse mWnt-medulloblastoma is functionally equivalent to mutation of the gene in human WNT-medulloblastoma, we compared single cell RNAseq (scRNAseq) profiles of DDX3X mutant (DDX3Xmut, n=2) and wild-type (DDX3Xwt, n=3) human WNT-medulloblastomas (Hovestadt et al., 2019). As expected, overall Uniform Manifold Approximation and Projection (UMAP) analysis of scRNAseq profiles segregated WNT-medulloblastoma cells according to the originating tumor/patient (Figure 6A). Therefore, to identify genes that might be dysregulated by mutation of DDX3X in WNT-medulloblastoma, we compared gene expression patterns in all cells derived from the two DDX3Xmut WNT-medulloblastomas (n=766 cells) with those in the three DDX3Xwt WNT-medulloblastomas (n=1,344 cells; Figure 6B). Remarkably, similar to our comparison of mWnt;Ddx3xFlx/Y and mWnt;Ddx3x+/+ mouse tumors (Figure 4B and C) genes upregulated in DDX3Xmut versus DDX3Xwt WNT-medulloblastoma cells were highly enriched for ‘stress granule associated’ (FDR=2.5e-38 - the most significantly upregulated specific cell function geneset), ‘stress response’ (FDR=2.2e-20), cell death/apoptosis (FDR=3.1e-8), and DNA repair genes (FDR=8.5e-8; Figure 6B). Several of these genes were commonly upregulated in both mouse mWNT;Ddx3xFlx/Y and human DDX3Xmut WNT-medulloblastoma and are implicated in other neurological disorders, including (Figure 4B, 6B): ANXA11 that regulates stress granule transport and is disrupted in amyotrophic lateral sclerosis (Liao et al., 2019); EPCAM, DLX1, FOXB1, IGFBP3, SLIT3, DDX20 that mediate cell death and apoptosis, particularly in neuronal tissues (Dai et al., 2017; Forones et al., 2018; Weber et al., 2018; Zhang et al., 2006); and the DNA repair enzymes RAD51 and RAD54L that are critically upregulated in other brain tumors (McKenzie et al., 2019b; Tong et al., 2015). Intriguingly, human DDX3Xmut WNT-medulloblastoma cells also upregulated cerebellar granule NPC markers, including NEUROD1, PAX6, ATAD2 and BAZ1A that were upregulated in mWnt;Ddx3xFlx/Y mouse cerebellar tumors (FDR=1.0e-4; Figure 4B and 6B). Thus, as suggested by recent imaging of human tumors (Di Giannatale et al., 2016), a small fraction of human DDX3Xmut WNT-medulloblastomas might arise within the cerebellar granule NPC lineage.
Figure 6. Single cell RNAseq of human DDX3Xmut and DDX3Xwt WNT-medulloblastoma.

A. UMAP of single cell RNAseq profiles generated from human DDX3Xmut (n=2) and DDX3Xwt (n=3) WNT-medulloblastoma. B. Volcano plots of -log10 p-values versus log fold difference expression of genes in the human DDX3Xmut and DDX3Xwt WNT-medulloblastomas in (A). Coloured genes in each plot belong to the cell pathway with associated FDR enrichment (top left). Exemplar genes are indicated; those marked with an asterix were also upregulated in mouse Blpb-Cre+/−;mWnt;Ddx3xFlx/Y cerebella tumors. See also Figure 4C and S5.
Finally, analysis of bulk RNAseq profiles generated from a separate cohort of human DDX3Xmut (n=14) and DDX3Xwt (n=14) WNT-medulloblastoma, identified 431 of 15,495 human-mouse orthologs that were upregulated in human mutDDX3X vs. wtDDX3X WNT-medulloblastoma (Figure S5). Ten percent (42/431) of these genes were also upregulated in mWnt;Ddx3xFlx/Y tumors (overlap representation factor=1.4, p=0.01) and were enriched for regulators of the cell stress (FDR=0.04 to 0.009) and immune response (FDR=0.03 to 1.2e−4), including: CCL2, SPINT1, SERTAD4, IFIT3, IFI44L, LRRC15, GCH1, DUSP9, RSAD2, and SELL implicated in inflammasome and innate immune signaling (Khan et al., 2015; Kitajima et al., 2018; Mathias et al., 2007; McNeill et al., 2018; O’Prey et al., 2008; Unamuno et al., 2019); and TRPV4 and NEXN that are implicated in neural inflammation (Khsheibun et al., 2014; Ye et al., 2012). Together, these data support the notion that DDX3X mediates a tumor suppressing, stress/inflammasome response in both human and mouse WNT-medulloblastoma that is partially relieved by loss of gene function.
DISCUSSION
DDX3X regulates an array of cell processes ranging from gene transcription and translation to the stress response (Herdy et al., 2018; Oh et al., 2016; Samir et al., 2019; Soto-Rifo et al., 2012; Valentin-Vega et al., 2016). Medulloblastoma-associated mutations in DDX3X have been shown to sequester DDX3X in stress granules and disrupt global patterns of gene translation (Oh et al., 2016; Valentin-Vega et al., 2016). But these studies have been conducted in vitro; therefore, it was unclear if DDX3X operates as a medulloblastoma oncogene or tumor suppressor gene. Here, we provide evidence that DDX3X serves as a key regulator of normal and malignant hindbrain development, suppressing WNT and SHH-medulloblastomas and restricting the competence of cell lineages to form these tumors.
Heterozygous, germline, missense mutations that disrupt DDX3X RNA helicase function account for up to 3% of intellectual disability in human females and cause hindbrain developmental defects very similar to those that we observed in female Blbp-Cre+/−;Ddx3xFlx/Flx mice (Dikow et al., 2017; Lennox et al., 2020; Nicola et al., 2019; Scala et al., 2019; Snijders Blok et al., 2015). Thus, Ddx3x deletion from the mouse brain phenocopies DDX3X loss-of-function mutations in humans. In our mice, aberrant hindbrain development was preceded by a highly-selective downregulation of Hox gene expression; suggesting strongly that Ddx3x patterns the hindbrain by regulating the transcription or stability of Hox RNAs. The transcription, splicing and stability of RNAs, including Hox transcripts, is critically dependent on guanine (G)-rich secondary structures in RNA termed G-quadruplexes (rG4) of which DDX3X is a key component (Herdy et al., 2018; Kwok et al., 2018; Kwok et al., 2016; Millevoi et al., 2012). Disruption of normal rG4s has been implicated in the pathogenesis of neurodegenerative diseases (Fay et al., 2017). Therefore, DDX3X might regulate Hox expression by maintaining transcript-associated rG4s. G4s also form within chromatin regulatory regions of DNA (dG4s) to control the transcription of Hox and other genes, and disruption of these structures has been implicated in cancer and neurodegeneration (Hansel-Hertsch et al., 2016; Lago et al., 2017; Zhang et al., 2018); therefore DDX3X might also regulate Hox-associated dG4s. Since Hox expression and brain development was largely unaffected in male Blbp-Cre+/−;Ddx3xFlx/Y mice, we propose that the paralog Ddx3y substitutes for Ddx3x during neurodevelopment, perhaps by substituting in rG4 and dG4s. This notion is consistent with the rarity of intellectual problems in human males with germline mutations in DDX3X and evidence that Ddx3y regulates NPC differentiation in vitro (Vakilian et al., 2015).
Our finding that Blbp-Cre+/−;Ddx3xFlx/Flx hindbrains also dysregulate cell stress, death and immune responses, unmasks a second potential function of DDX3X in normal and malignant hindbrain development –regulation of stress granule-inflammasome signaling. Ddx3x is a critical component of stress granules and inflammasomes (Samir et al., 2019; Shih et al., 2012). These multi-protein heteromeric complexes orchestrate the cell stress response and have been implicated in the pathogenesis of neurodegeneration and cancer (Anderson et al., 2015; Gan et al., 2018; Ivanov et al., 2019; Karki and Kanneganti, 2019; McKenzie et al., 2019a; Protter and Parker, 2016; Samir et al., 2019; Voet et al., 2019). Recently, we showed that Ddx3x shuttles between stress granules and inflammasomes to regulate cell fate choice in response to stress (Samir et al., 2019): incorporation of Ddx3x into stress granules promotes a cell survival signal, while its inclusion in inflammasomes drives pyroptosis - a form of programmed cell death important in the innate immune response and neurodegeneration (McKenzie et al., 2019a; Voet et al., 2019). Cre-recombination of Ddx3xFlx/Flx prevents Ddx3x incorporation in inflammasomes, blocking pyroptotic cell death signals and driving a default survival signal (Samir et al., 2019). Thus, disordered Blbp-Cre+/−;Ddx3xFlx/Flx hindbrain development might be exacerbated postnatally by the failure of pyroptotic cell death to remove abnormal cells. Although Ddx3y does not restore the pyroptotic response in Ddx3xFlx/Y recombined cells (Samir et al., 2019), Blbp-Cre+/−;Ddx3xFlx/Y hindbrains developed relatively normally and did not display aberrant cell stress, immune or death responses. Thus, analogous to the triggering of stress granule-inflammasome responses by amyloid-beta plaques in the brains of patients with Alzheimer’s disease (McKenzie et al., 2019a), abnormal hindbrain patterning in Blbp-Cre+/−;Ddx3xFlx/Flx mice - a feature absent from Blbp-Cre+/−;Ddx3xFlx/Y hindbrains - might trigger stress signals to which these mice are unable to respond appropriately (Figure 7).
Figure 7. Cartoon depicting the proposed role of Ddx3x in sensing oncogenic stress signals in the hindbrain.

In the context of wild-type Ddx3x shown left, oncogenic Wnt and Shh-signals in the lower and upper rhombic lip, respectively are relatively well tolerated. This provokes only a modest oncogenic stress response and fails to trigger a major inflammasome-pyroptotic death signal, markedly increasing the probability of forming medulloblastoma. Conversely, activation of oncogenic Wnt and Shh-signals in the upper and lower rhombic lip, respectively in which they are not primary mitogens, provokes a massive pyroptotic and tumor suppressing response. In the context of mutant Ddx3x shown right, this tumor suppressing pyroptotic cell death response is lost, allowing Wnt and Shh-medulloblastomas to form in either the upper or lower rhombic lips.
Our data also provide strong evidence that DDX3X functions as a medulloblastoma tumor suppressor gene: a notion compatible with studies of >4,100 cancers, including medulloblastoma, that identified DDX3X as an ‘escape from X-inactivation tumor-suppressor’ or EXIST gene (Dunford et al., 2017). Thus, male medulloblastomas harboring missense mutations that inactivate gene function do not require deletion of a second allele (Lennox et al., 2020). In female medulloblastoma, DDX3X might function as a haploinsufficient tumor suppressor gene or the remaining wild-type allele could undergo X-inactivation since this process is variable and context dependent (Garieri et al., 2018).
Particularly remarkable was our observation that deletion of Ddx3x increased the penetrance of both mWnt and mShh-medulloblastomas and derestricted the cell lineages able to form these tumors. We therefore provide an explanation for the frequent mutation of DDX3X in both of these tumor subtypes in humans, and insight into the mechanism(s) that establish medulloblastoma subtypes. Specifically, our RNAseq and immunohistochemical studies of premalignant cerebella showed that aberrant Wnt signaling in cerebellar granule NPCs provokes a potent oncogenic-stress response that was removed partially by the deletion of Ddx3x. Aberrant Wnt-, rather than Shh-signalling, provoked this profound stress response in cerebellar granule NPCs likely because the latter and not the former is a primary mitogen for this cell lineage. Therefore, we propose that DDX3X functions as a medulloblastoma tumor suppressor by activating inflammasome-mediated pyroptotic cell death in response to oncogenic stress (Figure 7). In doing so, DDX3X serves as a gatekeeper of WNT and SHH-medulloblastoma subtypes, removing SHH-mutant lower rhombic lip and WNT-mutant granule NPCs, contributing to the exquisite restriction of oncogenic WNT and SHH pathway mutations to these tumor types in upper and lower rhombic lip derived medulloblastomas, respectively. We are performing additional studies of Ddx3x suppression of mShh;Ddx3xFlx/Y and mShh;Ddx3xFlx/Flx brainstem tumorigenesis since limited tumor material precluded an equally thorough assessment of the origin of these tumors.
An important question raised by our work, is the extent to which DDX3X suppresses lineage origins of WNT and SHH-medulloblastomas in humans. In other words, what proportion of WNT and SHH-medulloblastomas, if any, arise from granule and lower rhombic lip NPCs, respectively, and do these harbor mutations in DDX3X? Answering this question will require meticulous correlation between pre-operative tumor imaging, inter-operative observation and tumor transcriptome and mutational landscape mapping. But existing imaging data suggest a proportion of WNT and SHH-medulloblastomas might arise from cerebellar granule and lower rhombic lip mossy fiber NPCs, respectively. The great majority of WNT-medulloblastomas are located in and around the cerebellar pontine angle and infiltrate the brainstem (Patay et al., 2015; Perreault et al., 2014); however, around 10% of cases do not involve the brainstem, with rare WNT-medulloblastomas being confined solely to the cerebellum (Di Giannatale et al., 2016). Similarly, while the majority of SHH-medulloblastomas are lateralized within the cerebellar hemispheres, a small proportion are found outside of the cerebellum, with rare cases being confined to the brainstem (Demir et al., 2019). It is noteworthy that patient age, tumor location and DDX3X mutational status are tightly correlated among human SHH-medulloblastomas: only 2% of SHH-medulloblastomas arising in patients aged ≤18 years mutate DDX3X and the majority are found in the cerebellar midline; conversely, 52% of adult SHH-medulloblastomas mutate DDX3X and almost all these tumors are located within the cerebellar hemispheres. Thus, adult SHH-medulloblastoma might arise within a distinct neural lineage in which DDX3X is particularly proficient at suppressing oncogenic signals.
It is important to note that while our mouse models delete Ddx3x, human medulloblastomas contain mainly heterozygous missense mutations in the gene (Northcott et al., 2019). However, evidence suggests that these alterations are likely to be functionally equivalent: developmental defects in Blbp-Cre+/−;Ddx3xFlx/Flx mice are strikingly similar to those seen in humans with de novo, heterozygous, germline DDX3X mutations; several missense mutations observed in humans with developmental defects are identical to those observed in medulloblastoma; and these mutations have been shown to impair RNA helicase activity and be functionally equivalent to knock-out of Ddx3x in the developing forebrain (Dikow et al., 2017; Lennox et al., 2020; Nicola et al., 2019; Scala et al., 2019; Snijders Blok et al., 2015). Furthermore, a common feature of DDX3X mutations associated with both developmental defects and medulloblastoma, is their capacity to sequester DDX3X within stress granules: the greater the degree of sequestration, the more severe the phenotype (Lennox et al., 2020; Valentin-Vega et al., 2016). Diversion of DDX3X into stress granules and away from inflammasomes would block pyroptotic cell death, resulting in a default cell survival signal and persistence of DDX3X-mutant tumor cells (Figure 7).
Finally, the discovery that DDX3X signaling suppresses both WNT and SHH-medulloblastomas opens up potential new avenues to treat these tumors. Extensive drug discovery programs yielding both DDX3X agonists and antagonists have been stimulated by evidence that DDX3X is a component of critical host-cell pathways, hijacked by several pathogenic human viruses (Brai et al., 2019; Valiente-Echeverría et al., 2015). We are currently working with groups studying infectious diseases to determine if DDX3X agonists might be used to restore gene function and pyroptotic response in medulloblastomas in which the gene is mutated.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Richard Gilbertson (Richard.gilbertson@cruk.cam.ac.uk).
Materials Availability
The Ddx3xFlx mouse line used in this study was generated by the authors and described previously. The line is available freely from the author and is in the process of being deposited with Jax. Mouse medulloblastoma cells generated from mice that are described in this study are available freely from the lead contact under a standard Materials Transfer Agreement. Only costs to cover post and packaging will be requested.
Data and Code Availability
The RNA and DNA sequencing datasets generated during this study have been submitted to the GEO database (Accession ID: GSE147178 for RNA-sequencing and GSE147069 for DNA-sequencing). Other RNA sequencing sets that were previously published are referenced and available as indicated in the respective papers.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Genetically Engineered Models
All animal studies were performed under the Animals (Scientific Procedures) Act 1986 in accordance with UK Home Office licenses (Project License 70–8798, P6787677F) and approved by the Cancer Research UK (CRUK) Cambridge Institute Animal Welfare and Ethical Review Board. Mice were maintained in conventional cages and given access to standard diet and water. The developmental stage, sex and number of animals in each experiment is reported directly in the main Results section of the manuscript. Since our studies focused on the X-linked gene Ddx3x, then mice were not randomly assigned by sex to study groups. Control groups included an equal number of relevant sex animals unless indicated.
Generation of the Ddx3x conditional allele (Ddx3xFlx) was detailed previously (Samir et al., 2019). Briefly, the Ddx3x allele was modified using CRISPR-Cas9 gene editing. Pronuclear stage zygotes were injected with two single guide RNAs targeting intron 6 and 14 of Ddx3x, Cas9 mRNA transcripts and homology directed repair oligonucleotides (Ddx3x-Intron6-LoxP-HDR and Ddx3x-Intron14-LoxP-HDR) designed to incorporate loxP sites together with EcoRI restriction sites to facilitate targeting and genotyping. Mice carrying the Ddx3xFlx allele were then bred with those harbouring the Blbp-Cre allele resulting in deletion of exons to 7–14 of the recombined Ddx3xFlx allele. mWnt mice were described previously (Gibson et. al., 2010). mShhmedulloblastomas were generated by breeding mice carrying the Blbp-Cre+/− allele with B6N.129-Ptch1tm1Hahn/J mice (The Jackson Laboratory, 012457).
Orthotopic allografting of mouse tumors
Mouse tumors were allografted into 4–6 week old female CD1-Foxn1NU mice (Charles River, 086) exactly as described previously (Mohankumar et al., 2015). Briefly, 5×105 cells transduced with a YFP-luciferase lentivirus were re-suspended in 5μl matrigel (Corning, 354230) and injected via the cistern magna into the cerebellum of anaesthetised CD1-Foxn1NU mice. Mice were monitored daily for neurological symptoms and weekly using bioluminescence imaging to detect tumor growth.
METHOD DETAILS
Western Blot Analysis
Cell pellets were harvested and lysed using RIPA lysis buffer (Invitrogen, 89901). 20μg of protein derived from relevant cell pellets were loaded into each lane of a standard SDS-PAGE gel and resolved by electrophoresis (Thermo Fisher Scientific, NP0335). Following transfer to 0.2μm nitrocellulose membrane (Thermo Fisher Scientific, IB301001), membranes were exposed to primary antibodies directed against Ddx3x (Bethyl, A300–474A) and beta-actin (Cell Signaling, 4967) and 1:15,000 dilution of anti-rabbit IRDye680LT-conjugated secondary antibody (LI-COR, 926–68023). Antibody binding to proteins was detected using the LI-COR Odyssey CLx.
Histology and Analysis
Tissue samples were formalin fixed, paraffin embedded and cut into 5μm sections as described (Gibson et al., 2010). Hematoxylin and Eosin (H&E) staining (Thermo Fisher Scientific, 7221 and 7111), was performed according to manufacturer’s instructions. For the nissl staining, sections were stained with 0.5% cresyl fast violet (Sigma-Aldrich, H3757) for 10 minutes, washed with ultra-pure water followed by 0.25% acetic acid (Sigma-Aldrich, A6283) to clear the background stain. Sections were then dehydrated through serial alcohol solutions, rinsed in xylene and cover slipped. Immunohistochemistry was performed using the Leica Polymer Refine Detection System (Leica Biosystems, DS9800) together with the automated Bond platform. Primary antibodies included: Ki67 (1:1000; Bethyl, IHC-00375), cleaved caspase 3 (1:200; Cell Signaling, 9664), p21 (1:100; BD Biosciences, 556431), CD3 (Agilent Dako, A0452), CD45 (Abcam, ab25386), CD68 (Abcam, ab53444) and F4/80 (Bio-Rad, MCA497). Secondary (post primary) antibodies were anti-rabbit Poly-HRP-IgG (included in kit) and rabbit anti-mouse IgG1 (1:1500; Abcam, ab6728). Sections were then dehydrated through serial alcohol solutions, rinsed in xylene and cover slipped. Digital images of entire tissue sections were generated using the Leica Aperio AT2 digital scanner. Images were captured at 20x magnification, with a resolution of 0.5uM/pixel. Images were then viewed using the Leica Aperio Image Scope v12.3.2.8013 and staining quantified automatically using the Aperio ePathology image analysis software.
Immunofluorescence Staining
Immunofluorescence was performed using sections of formalin fixed, paraffin embedded tissue generated as described above. Antigen retrieval in tissue sections was achieved using pressure-cooking in citrate buffer pH6 for 20 minutes. Tissue sections were incubated with primary antibodies for 1 hour at room temperature.
Primary antibodies included: Ki67 (1:200; Abcam, ab15580 and 1:500; Thermo Fisher Scientific, 14-5698-82), Calbindin (1:50; Millipore, AB1778), Contactin2 (1:75; R&D Systems, AF4439), Lef1 (1:500; Abcam, ab137872), Pax6 (1:200; Biolegend, 901301), G3BP1 (1:100; Proteintech, 66486-1-Ig), Gfap (1:500; Dako, Z0334), NeuN (1:200; Millipore, MAB377), Irf1 (1:100; Abcam, ab240299), Iba1 (1:500; Wako, 013-27691) and Synaptophysin (1:100; Cell Signaling, 9020). Following washing, tissue sections where then incubated for 1 hour at room temperature in secondary antibody. Secondary antibodies included Alexa 488, 594 or 680 (1:200; Invitrogen, A-21206, A-21207 or A10043). Sections were then counterstained using DAPI (Cell Signaling, 4083) and mounted using ProLong Gold antifade mountant (Thermo Fisher Scientific, P36930). Digital images of tissue sections were captured using a Zeiss ImagerM2 and Apotome microscope.
In Situ Hybridisation
Ddx3x RNA expression was detected in formalin fixed paraffin embedded tissue sections simultaneously with mouse Gbx2, Otx2, Pax5, Atoh1 or Hoxa2 (see probe details in STAR Methods Table) using the Advanced Cell Diagnostics (ACD) RNAscope® 2.5 LS Duplex Reagent Kit (ACD, 322440). Briefly, 5μM thick, freshly cut sections were baked for 1 hour at 60°C and then loaded onto a Bond RX instrument (Leica Biosystems). Slides were deparaffinized and rehydrated on board the Bond RX before pre-treatment with Epitope Retrieval Solution 2 (Leica Biosystems, AR9640,) at 95°C for 10 minutes, and ACD Enzyme (Duplex Reagent kit) at 40°C for 15 minutes. Probe hybridisation and signal amplification was performed according to manufacturer’s instructions. Fast red detection of mouse Gbx2, Otx2, Pax5, Atoh1, or Hoxa2 was performed on the Bond Rx using the Bond Polymer Refine Red Detection Kit (Leica Biosystems, DS9390) according to manufacturer’s protocol. Slides were then removed from the Bond Rx and detection of Ddx3x RNA performed using the RNAscope® 2.5 LS Green Accessory Pack (ACD, 322550) according to kit instructions. Slides were heated at 60°C for 1 hour, dipped in xylene and mounted using VectaMount permanent mounting medium (Vector Laboratories, H-5000). The slides were imaged on the Aperio AT2 (Leica Biosystems) to create whole slide images. Images were captured at 40x magnification, with a resolution of 0.25 microns per pixel.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Donkey anti-Rabbit IgG, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21206; RRID:AB 2535792 |
| Donkey anti-Rabbit IgG, Alexa Fluor 564 | Thermo Fisher Scientific | Cat# A-21207; RRID:AB 141637 |
| Donkey anti-Rabbit IgG, Alexa Fluor 680 | Thermo Fisher Scientific | Cat# A10043; RRID;AB 2534018 |
| Goat polyclonal anti-Contactin2 | R&D Systems | Cat# AF4439; RRID:AB 2044647 |
| Mouse monoclonal anti-G3BP1 | Proteintech | Cat# 66486-1-Ig; RRID:AB 2819031 |
| Mouse monoclonal anti-NeuN | Millipore | Cat # MAB377; RRID:AB 2298772 |
| Mouse monoclonal anti-p21 | BD Biosciences | Cat# 556431; RRID:AB 396415 |
| Mouse monoclonal anti- Synaptophysin | Cell Signaling | Cat# 9020; RRID:AB_2631095 |
| Rabbit monoclonal anti-Cleaved Caspase-3 | Cell Signaling | Cat# 9664; RRID:AB_2070042 |
| Rabbit monoclonal anti-Irf1 | Abcam | Cat# ab240299, RRID:AB 2737059 |
| Rabbit monoclonal anti-Lef1 | Abcam | Cat# ab137872; RRID:AB 1267233 |
| Rabbit monoclonal anti-Mouse IgG | Abcam | Cat# ab6728; RRID:AB 955440 |
| Rabbit polyclonal anti-Beta actin | Cell Signaling | Cat# 4967; RRID:AB 330288 |
| Rabbit polyclonal anti-Calbindin | Millipore | Cat# AB1778; RRID:AB 2068336 |
| Rabbit polyclonal anti-CD3 | Agilent Dako | Cat# A0452, RRID:AB 2335677 |
| Rabbit polyclonal anti-Ddx3x | Bethyl | Cat# A300–474A; RRID:AB 451009 |
| Rabbit polyclonal anti-Gfap | Agilent Dako | Cat# Z0334; RRID:AB 10013382 |
| Rabbit polyclonal anti-Iba1 | Wako | Cat# 013–27691, RRID:AB 839504 |
| Rabbit polyclonal anti-Ki67 | Abcam | Cat# ab15580; RRID:AB 443209 |
| Rabbit polyclonal anti-Ki67 | Bethyl | Cat# IHC-00375; RRID:AB 1547959 |
| Rabbit polyclonal anti-Pax-6 | Biolegend | Cat# 901301; RRID:AB 2565003 |
| Rat monoclonal anti-CD45 | Abcam | Cat# ab25386, RRID:AB 470499 |
| Rat monoclonal anti-CD68 | Abcam | Cat# ab53444, RRID:AB 869007 |
| Rat monoclonal anti-F4/80 | Bio-Rad | Cat# MCA497, RRID:AB 2098196 |
| Rat monoclonal anti-Ki67 | Thermo Fisher Scientific | Cat# 14-5698-82; RRID:AB 10854564 |
| IRDye 680LT Donkey anti rabbit IgG | LI-COR | Cat# 926-68023; RRID:AB 10706167 |
| Bacterial and Virus Strains | ||
| YFP-luciferase lentivirus | Mohankumar et al., 2015 | NA |
| Cre-recombinase lentivirus | Tong et. al., 2015 | NA |
| Biological Samples | ||
| Mouse Cerebellum | This manuscript | NA |
| Mouse MB Tumours | This manuscript | NA |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Accutase | Thermo Fisher Scientific | Cat# A1110501 |
| Acetic Acid | Sigma-Aldrich | Cat# A6283 |
| B-27 Supplement | Thermo Fisher Scientific | Cat# 12587010 |
| Collagenase Type IV | Thermo Fisher Scientific | Cat# 17104-019 |
| Cresyl Fast Violet | Sigma-Aldrich | Cat# C5042 |
| DAPI | Cell Signaling | Cat# 4083 |
| Epitope Retrieval Solution 2 | Leica Biosystems | Cat# AR9640 |
| Eosin | Thermo Fisher Scientific | Cat# 7111 |
| Hematoxylin | Thermo Fisher Scientific | Cat# 7221 |
| Human EGF | MACS Miltenyi Biotec | Cat# 130-097-751 |
| Human FGF-2 | MACS Miltenyi Biotec | Cat# 130-093-843 |
| Hyaluronidase | Sigma-Aldrich | Cat# H3757 |
| iBlot™ Transfer Stack, nitrocellulose | Thermo Fisher Scientific | Cat# IB301001 |
| Matrigel Matrix | Corning | Cat# 354230 |
| N-2 Supplement | Thermo Fisher Scientific | Cat# A1370701 |
| Neurobasal Medium | Thermo Fisher Scientific | Cat# 21103049 |
| Normal Donkey Serum | Sigma-Aldrich | Cat# D9663 |
| NuPAGE, 4–12% Bis-Tris protein gel | Thermo Fisher Scientific | Cat# NP0335 |
| ProLong Gold antifade mountant | Thermo Fisher Scientific | Cat# P36930 |
| Protease Inhibitors | Sigma-Aldrich | Cat# P8340 |
| RIPA Lysis Buffer | Invitrogen | Cat# 89901 |
| RNAscope® 2.5 LS Probe - Mm- Ddx3x | Advanced Cell Diagnostics | Cat# 490438 |
| RNAscope® 2.5 LS Probe- Mm- Atoh1 | Advanced Cell Diagnostics | Cat# 408798 |
| RNAscope® 2.5 LS Probe- Mm- Gbx2 | Advanced Cell Diagnostics | Cat# 314358 |
| RNAscope® 2.5 LS Probe- Mm- Hoxa2 | Advanced Cell Diagnostics | Cat# 451268 |
| RNAscope® 2.5 LS Probe- Mm- Otx2 | Advanced Cell Diagnostics | Cat# 444388 |
| RNAscope® 2.5 LS Probe- Mm- Pax5 | Advanced Cell Diagnostics | Cat# 311488 |
| VectaMount Permanent Mounting Medium | Vector Laboratories | Cat# H-5000 |
| Critical Commercial Assays | ||
| Kapa qPCR library quantification kit | Kapa Biosystems | Cat# KK4873 |
| TruSeq stranded mRNA kit | Illumina | Cat# 20020595 |
| Leica Polymer Refine Detection System | Leica Biosystems | Cat# DS9800 |
| Nextera Flex pre-enrichment kit | Illumina | Cat# 20025523 |
| Polymer Refine Red Detection Kit | Leica Biosystems | Cat# DS9390 |
| RNAscope® 2.5 LS Duplex Reagent Kit | Advanced Cell Diagnostics | Cat# 3224400 |
| RNAscope® 2.5 LS Green Accessory Pack | Advanced Cell Diagnostics | Cat# 322550 |
| RNeasy Plus universal kit | Qiagen | Cat# 73404 |
| truXTRAC FFPE Total NA Plus Kit | Covaris | Cat# PN 520255 |
| Deposited Data | ||
| DNA sequencing data | This manuscript | GSE147069 |
| RNA sequencing data | This manuscript | GSE147178 |
| Experimental Models: Cell Lines | ||
| Mouse: Ddx3xflx | This manuscript | NA |
| Mouse: Blbp-Cre+/−;Ctnnb 1 +/lox(ex3);Tp53 +/flx | This manuscript | NA |
| Mouse: Blbp-Cre+/−;Ddx3xtlx/tlx;Ctnnb 1 +/lox(ex3) ;Tp53 +/flx | This manuscript | NA |
| Experimental Models: Organisms/Strains | ||
| Ddx3xflx | Samir et. al. 2019 | NA |
| Blbp-Cre+/−; Ctnnb1+/lox(ex3);Tp53+/tlx | Gibson et. al. 2010 | NA |
| Blbp-Cre+/−; Ddx3xflx; Ctnnb1+/lox(ex3); Tp53+/flx | This manuscript | NA |
| Ptch1tm1Hahn (Ptch+/−) | The Jackson Laboratory; Uhmann et. al. 2007 | Stock# 012457 (RRID: IMSR_JAX:012457) |
| Blbp-Cre+/−; Ddx3xflx; Ptch+/− | This manuscript | NA |
| CD1-Foxn1NU | Charles River | Strain # 086; RRID:IMSR_CRL:086 |
| Oligonucleotides | ||
| Ddx3x Intron 6 forward primer | Integrated DNA Technologies | TTCTGGAGGCAATACTGGGA |
| Ddx3x Intron 6 reverse primer | Integrated DNA Technologies | ATGAGCTTACCTGTTTGAGCA |
| Ddx3x Intron 14 forward primer | Integrated DNA Technologies | CCGTGTGGGAAACCTTGGTA |
| Ddx3x Intron 14 reverse primer | Integrated DNA Technologies | AGAACGTCCACGACTGCTAC |
| Ptch forward primer | Integrated DNA Technologies | TTCATTGAACCTTGGGGAAC |
| Ptch reverse primer | Integrated DNA Technologies | AGTGCGTGACACAGATCAGC |
| Recombinant DNA | ||
| Software and Algorithms | ||
| Aperio ePathology | Leica Biosystems | www.leicabiosystems.com |
| Aperio ImageScope | Leica Biosystems | www.leicabiosystems.com |
| Burrows-Wheeler Aligner (BWA) | (Li and Durbin, 2010) | www.sourceforge.net/projects/bio-bwa/ |
| CGHcall package (version 2.44.0) | (van de Wiel et al., 2007) | www.bioconductor.org/packages/release/bioc/html/CGHcall |
| DESeq2 package (version 1.26.0) | (Love et al., 2014) | www.bioconductor.org/packages/release/bioc/html/DESea2 |
| DNAcopy package (version 1.56.0) | R package version 1.58.0 | www.bioconductor.org/packages/release/bioc/html/DNAcopy |
| GraphPad Prism 6.00 | GraphPad | www.graphpad.com |
| QDNAseq package (version 1.12.0) | (Scheinin et al., 2014) | www.bioconductor.org/packages/release/bioc/html/QDNAsea |
| Other | ||
| Ultra-low attachment T75 flask | Corning | Cat# 3814 |
RNA and DNA Extractions
Total RNA was extracted from tissue material using the RNeasy Plus universal kit (Qiagen, 73404) and stored at −80° C. DNA was extracted from 30μm thick formalin fixed, paraffin tissue sections using the truXTRAC FFPE total NA Plus Kit (Covaris, 520255) followed by magnetic bead-based DNA purification. DNA quality was verified using Tapestation (Agilent).
Library Preparation and Sequencing
All library preparation and sequencing were performed by the Genomics Core at the CRUK Cambridge Institute. The Illumina TruSeq stranded mRNA kit (Illumina, 20020595) was used to prepare RNA libraries and RNA quality confirmed using Tapestation (Agilent) and quantified using Kapa qPCR library quantification kit for Illumina platforms (Kapa Biosystems, KK4873). Samples were normalised using the Agilent Bravo, pooled and sequenced on Illumina HiSeq 4000 to generate single end 50bp reads at 20M reads per sample. DNA sequencing was performed on DNA libraries prepared using Nextera Flex preenrichment kit (Illumina) and sequenced on a NovaSeq SP flowcell generating paired end 50 reads at 20–42M reads per sample.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantitative histological analysis
Nuclear immunoreactivity for tested antigens was quantified using the nuclear v9 macro/algorithm (H&E, Ki67 and p21) and cytoplasmic immunoreactivity using the cytoplasmic v2 macro/algorithm (cleaved caspase 3). The identical pipeline was used for all tissue sections. The percentage of positive cells in each tissue section identified using these algorithms was recorded relative to the total number of nuclei in each section. Details of the numbers of tissues employed in each analysis are reported directly in the relevant part of the text of the main Results section.
Analysis of RNA sequence data
Single end 50bp RNA reads were aligned to GRCm38 with HISAT2 (with default parameters), each individual BAM file sorted and annotated. Each sample was sequenced across several lanes; per-lane BAM files were merged into per-sample BAMs. Quality control metrics were collected for each file, including duplication statistics and number of reads assigned to genes. Reads were counted on annotated features with sub-reads featureCounts, providing ‘total’, ‘aligned to the genome’, and ‘assigned to a gene’ (i.e. included in the analysis) counts. Percentages of aligned bases were computed for several categories: coding, UTR, intronic, intergenic. Other quality control metrics were the percentage of reads on the correct strand, median coefficient of variation of coverage, median 5’ bias, median 3’ bias, and the ratio of 5’ to 3’ coverage. Quality control also included an expression heatmap drawn using log2 transformed counts. Log2 transformed counts were generated from normalized counts using the log2 function in R and counts function from DEseq2. Genes were regarded as displaying differential expression between sample cohorts if they displayed of ≥1 or ≤−1 log-fold differences in expression levels and an absolute difference in mean gene expression level between groups of >50. Differential Principal Component Analysis was carried out on variance-stabilizing-transformed raw counts using the ‘vst’ function from DESeq2. Details of the numbers of tissues employed in each analysis are reported directly in the relevant part of the text of the main Results section.
Normalised TPM counts of WNT-medulloblastoma single cell RNA sequencing were log-transformed and then passed into a Seurat object. Cells were filtered based on a minimum of 200 and maximum of 9000 genes per cell. Normalized counts were then passed directly to the FindVariableFeatures and ScaleData functions from the Seurat package. Effects from total counts of RNA were regressed out in the ScaleData function. Downstream clustering analysis was then carried out in the Seurat package using default parameters. Details of the numbers of tissues employed in each analysis are reported directly in the relevant part of the text of the main Results section.
Pathway analysis
Genes identified as up- or downregulated within bulk and single cell RNAseq profiles between different tissue and cell types were analyzed using g:GOST within the g:Profiler interface (Raudvere et al., 2019). g:GOST enabled identification of statistically enriched genes that over-representated Gene Ontology terms, biological pathways, regulatory DNA elements, human disease gene annotations, and protein-protein interaction networks. Visualization of patterns of pathway enrichment in g:GOST outputs were generated using Enrichment map within Cytoscape (Shannon et al., 2003).
DNA copy number analysis
Raw DNA sequence reads were aligned to the mm10 assembly of the mouse genome (mm10, Genome Reference Consortium Mouse Build 38 (GCA_000001635.2)) from UCSC. Read mapping was performed using the Burrows-Wheeler Aligner. Copy number variation analysis was performed in R (version 3.5.2) using the QDNAseq package (version 1.12.0) from Bioconductor. A bin size of 100kb was used for ‘windowing’ copy number. Segmentation of copy number was carried out using the DNAcopy package (version 1.56.0) and gains and losses were called using the CGHcall package (version 2.44.0).
Statistical tests and survival analysis
Survival analyses among animal cohorts of the genotype, number and sex defined in the main text was performed using the Log-Rank Mantel-Cox statistic. Statistical differences in tissue histology patterns e.g., number of immune-positive nuclei, among animal cohorts of the genotype, number and sex defined in the main text was performed using the non-parametric Mann-Whitney test. Details of the numbers of tissues employed in each analysis are reported directly in the relevant part of the text of the main Results section.
Supplementary Material
HIGHLIGHTS.
DDX3X is frequently mutated in the brain tumor medulloblastoma
Ddx3x regulates hindbrain development via Hox gene expression and stress signaling
Ddx3x supresses medulloblastoma by restricting tumor lineage origins
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (R.J.G., P01CA96832, R01CA129541 and P30CA021765), the American Lebanese Syrian Associated Charities and Cancer Research UK. We are grateful to the staff of the BRU, Histology, Genomics, Bioinformatics, and Light Microscopy Core Facilities at the CRUK Cambridge Institute for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
None
REFENCENCES
- Anderson P, Kedersha N, and Ivanov P (2015). Stress granules, P-bodies and cancer. Biochim Biophys Acta 1849, 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brai A, Martelli F, Riva V, Garbelli A, Fazi R, Zamperini C, Pollutri A, Falsitta L, Ronzini S, Maccari L, et al. (2019). DDX3X Helicase Inhibitors as a New Strategy To Fight the West Nile Virus Infection. J Med Chem 62, 2333–2347. [DOI] [PubMed] [Google Scholar]
- Dai H, Goto Y-I, and Itoh M (2017). Insulin-Like Growth Factor Binding Protein-3 Deficiency Leads to Behavior Impairment with Monoaminergic and Synaptic Dysfunction. Am J Pathol 187, 390–400. [DOI] [PubMed] [Google Scholar]
- Demir MK, Yapıcıer Ö, Mert B, Alshareefi W, and Bozbuğa M (2019). Primary Sonic Hedgehog-activated dorsal brainstem medulloblastoma and ipsilateral cerebellar atrophy in an adult. Neuroradiol J, 1971400919892824–1971400919892824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giannatale A, Carai A, Cacchione A, Marrazzo A, Dell’Anna VA, Colafati GS, Diomedi-Camassei F, Miele E, Po A, Ferretti E, et al. (2016). Anomalous vascularization in a Wnt medulloblastoma: a case report. BMC Neurol 16, 103–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikow N, Granzow M, Graul-Neumann LM, Karch S, Hinderhofer K, Paramasivam N, Behl L-J, Kaufmann L, Fischer C, Evers C, et al. (2017). DDX3X mutations in two girls with a phenotype overlapping Toriello-Carey syndrome. Am J Med Genet A 173, 1369–1373. [DOI] [PubMed] [Google Scholar]
- Doherty GJ, Petruzzelli M, Beddowes E, Ahmad SS, Caldas C, and Gilbertson RJ (2019). Cancer Treatment in the Genomic Era. Annual Review of Biochemistry 88, 247–280. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno-Jones S, and Sawyers CL (2001). Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med 344, 1031–1037. [DOI] [PubMed] [Google Scholar]
- Duijkers FA, McDonald A, Janssens GE, Lezzerini M, Jongejan A, van Koningsbruggen S, Leeuwenburgh-Pronk WG, Wlodarski MW, Moutton S, Tran-Mau-Them F, et al. (2019). HNRNPR Variants that Impair Homeobox Gene Expression Drive Developmental Disorders in Humans. Am J Hum Genet 104, 1040–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunford A, Weinstock DM, Savova V, Schumacher SE, Cleary JP, Yoda A, Sullivan TJ, Hess JM, Gimelbrant AA, Beroukhim R, et al. (2017). Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat Genet 49, 10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay MM, Anderson PJ, and Ivanov P (2017). ALS/FTD-Associated C9ORF72 Repeat RNA Promotes Phase Transitions In Vitro and in Cells. Cell Rep 21, 3573–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forones NM, Lima FT, Martin RP, Martins L, Teixeira PVL, Pesquero JB, Oshima CTF, and Pimenta CAM (2018). Mismatch Repair Genes and EPCAM germline mutations in patients with gastric or colorectal cancer with suspected of Lynch syndrome. Journal of Clinical Oncology 36, e13623–e13623. [Google Scholar]
- Gajjar A, Chintagumpala M, Ashley D, Kellie S, Kun LE, Merchant TE, Woo S, Wheeler G, Ahern V, Krasin MJ, et al. (2006). Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multicentre trial. The Lancet Oncology 7, 813–820. [DOI] [PubMed] [Google Scholar]
- Gan L, Cookson MR, Petrucelli L, and La Spada AR (2018). Converging pathways in neurodegeneration, from genetics to mechanisms. Nat Neurosci 21, 1300–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garieri M, Stamoulis G, Blanc X, Falconnet E, Ribaux P, Borel C, Santoni F, and Antonarakis SE (2018). Extensive cellular heterogeneity of X inactivation revealed by single-cell allele-specific expression in human fibroblasts. Proceedings of the National Academy of Sciences 115, 13015–13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson P, Tong Y, Robinson G, Thompson MC, Currle DS, Eden C, Kranenburg TA, Hogg T, Poppleton H, Martin J, et al. (2010). Subtypes of medulloblastoma have distinct developmental origins. Nature 468, 1095–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, Di Antonio M, Pike J, Kimura H, Narita M, et al. (2016). G-quadruplex structures mark human regulatory chromatin. Nat Genet 48, 1267–1272. [DOI] [PubMed] [Google Scholar]
- Herdy B, Mayer C, Varshney D, Marsico G, Murat P, Taylor C, D’Santos C, Tannahill D, and Balasubramanian S (2018). Analysis of NRAS RNA G-quadruplex binding proteins reveals DDX3X as a novel interactor of cellular G-quadruplex containing transcripts. Nucleic Acids Res 46, 11592–11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovestadt V, Smith KS, Bihannic L, Filbin MG, Shaw ML, Baumgartner A, DeWitt JC, Groves A, Mayr L, Weisman HR, et al. (2019). Resolving medulloblastoma cellular architecture by single-cell genomics. Nature 572, 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Kedersha N, and Anderson P (2019). Stress Granules and Processing Bodies in Translational Control. Cold Spring Harbor perspectives in biology 11, a032813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessa S, Blanchet-Cohen A, Krug B, Vladoiu M, Coutelier M, Faury D, Poreau B, De Jay N, Hébert S, Monlong J, et al. (2019). Stalled developmental programs at the root of pediatric brain tumors. Nat Genet 51, 1702–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DTW, Jager N, Kool M, Zichner T, Hutter B, Sultan M, Cho Y-J, Pugh TJ, Hovestadt V, Stutz AM, et al. (2012). Dissecting the genomic complexity underlying medulloblastoma. Nature 488, 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki R, and Kanneganti T-D (2019). Diverging inflammasome signals in tumorigenesis and potential targeting. Nature reviews Cancer 19, 197–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi D, Robinson G, Uziel T, Gibson P, Rehg J, Gao C, Finkelstein D, Qu C, Pounds S, Ellison David W., et al. (2012). A Mouse Model of the Most Aggressive Subgroup of Human Medulloblastoma. Cancer Cell 21, 168–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan KA, Dô F, Marineau A, Doyon P, Clément J-F, Woodgett JR, Doble BW, and Servant MJ (2015). Fine-Tuning of the RIG-I-Like Receptor/Interferon Regulatory Factor 3-Dependent Antiviral Innate Immune Response by the Glycogen Synthase Kinase 3/β-Catenin Pathway. Mol Cell Biol 35, 3029–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khsheibun R, Paperna T, Volkowich A, Lejbkowicz I, Avidan N, and Miller A (2014). Gene expression profiling of the response to interferon beta in Epstein-Barr-transformed and primary B cells of patients with multiple sclerosis. PLoS One 9, e102331–e102331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiecker C, and Lumsden A (2005). Compartments and their boundaries in vertebrate brain development. Nat Rev Neurosci 6, 553–564. [DOI] [PubMed] [Google Scholar]
- Kitajima S, Asahina H, Chen T, Guo S, Quiceno LG, Cavanaugh JD, Merlino AA, Tange S, Terai H, Kim JW, et al. (2018). Overcoming Resistance to Dual Innate Immune and MEK Inhibition Downstream of KRAS. Cancer Cell 34, 439–452.e436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok CK, Marsico G, and Balasubramanian S (2018). Detecting RNA G-Quadruplexes (rG4s) in the Transcriptome. Cold Spring Harbor Perspectives in Biology 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok CK, Marsico G, Sahakyan AB, Chambers VS, and Balasubramanian S (2016). rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat Methods 13, 841–844. [DOI] [PubMed] [Google Scholar]
- Lago S, Tosoni E, Nadai M, Palumbo M, and Richter SN (2017). The cellular protein nucleolin preferentially binds long-looped G-quadruplex nucleic acids. Biochim Biophys Acta Gen Subj 1861, 1371–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox AL, Hoye ML, Jiang R, Johnson-Kerner BL, Suit LA, Venkataramanan S, Sheehan CJ, Alsina FC, Fregeau B, Aldinger KA, et al. (2020). Pathogenic DDX3X Mutations Impair RNA Metabolism and Neurogenesis during Fetal Cortical Development. Neuron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letai A (2017). Functional precision cancer medicine—moving beyond pure genomics. Nature Medicine 23, 1028–1035. [DOI] [PubMed] [Google Scholar]
- Li H, and Durbin R (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y-C, Fernandopulle MS, Wang G, Choi H, Hao L, Drerup CM, Patel R, Qamar S, Nixon-Abell J, Shen Y, et al. (2019). RNA Granules Hitchhike on Lysosomes for Long-Distance Transport, Using Annexin A11 as a Molecular Tether. Cell 179, 147–164.e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makki N, and Capecchi MR (2011). Identification of novel Hoxa1 downstream targets regulating hindbrain, neural crest and inner ear development. Dev Biol 357, 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias JR, Dodd ME, Walters KB, Rhodes J, Kanki JP, Look AT, and Huttenlocher A (2007). Live imaging of chronic inflammation caused by mutation of zebrafish Hai1. J Cell Sci 120, 3372–3383. [DOI] [PubMed] [Google Scholar]
- McKenzie BA, Dixit VM, and Power C (2019a). Fiery Cell Death: Pyroptosis in the Central Nervous System. Trends Neurosci, S0166–2236(0119)30218–30218. [DOI] [PubMed] [Google Scholar]
- McKenzie LD, LeClair JW, Miller KN, Strong AD, Chan HL, Oates EL, Ligon KL, Brennan CW, and Chheda MG (2019b). CHD4 regulates the DNA damage response and RAD51 expression in glioblastoma. Sci Rep 9, 4444–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill E, Stylianou E, Crabtree MJ, Harrington-Kandt R, Kolb A-L, Diotallevi M, Hale AB, Bettencourt P, Tanner R, O’Shea MK, et al. (2018). Regulation of mycobacterial infection by macrophage Gch1 and tetrahydrobiopterin. Nature communications 9, 5409–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Moine H, and Vagner S (2012). G-quadruplexes in RNA biology. Wiley Interdiscip Rev RNA 3, 495–507. [DOI] [PubMed] [Google Scholar]
- Mohankumar KM, Currle DS, White E, Boulos N, Dapper J, Eden C, Nimmervoll B, Thiruvenkatam R, Connelly M, Kranenburg TA, et al. (2015). An in vivo screen identifies ependymoma oncogenes and tumor-suppressor genes. Nat Genet 47, 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola P, Blackburn PR, Rasmussen KJ, Bertsch NL, Klee EW, Hasadsri L, Pichurin PN, Rankin J, Raymond FL, Study DDD, and Clayton-Smith J (2019). De novo DDX3X missense variants in males appear viable and contribute to syndromic intellectual disability. Am J Med Genet A 179, 570–578. [DOI] [PubMed] [Google Scholar]
- Northcott PA, Jones DTW, Kool M, Robinson GW, Gilbertson RJ, Cho Y-J, Pomeroy SL, Korshunov A, Lichter P, Taylor MD, and Pfister SM (2012a). Medulloblastomics: the end of the beginning. Nat Rev Cancer 12, 818–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins CE, French P, et al. (2011). Medulloblastoma Comprises Four Distinct Molecular Variants. Journal of Clinical Oncology 29, 1408–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Robinson GW, Kratz CP, Mabbott DJ, Pomeroy SL, Clifford SC, Rutkowski S, Ellison DW, Malkin D, Taylor MD, et al. (2019). Medulloblastoma. Nature Reviews Disease Primers 5, 11. [DOI] [PubMed] [Google Scholar]
- Northcott PA, Shih DJH, Peacock J, Garzia L, Sorana Morrissy A, Zichner T, Stutz AM, Korshunov A, Reimand J, Schumacher SE, et al. (2012b). Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 488, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Prey J, Wilkinson S, and Ryan KM (2008). Tumor antigen LRRC15 impedes adenoviral infection: implications for virus-based cancer therapy. J Virol 82, 5933–5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Flynn RA, Floor SN, Purzner J, Martin L, Do BT, Schubert S, Vaka D, Morrissy S, Li Y, et al. (2016). Medulloblastoma-associated DDX3 variant selectively alters the translational response to stress. Oncotarget 7, 28169–28182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patay Z, DeSain LA, Hwang SN, Coan A, Li Y, and Ellison DW (2015). MR Imaging Characteristics of Wingless-Type-Subgroup Pediatric Medulloblastoma. AJNR Am J Neuroradiol 36, 2386–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault S, Ramaswamy V, Achrol AS, Chao K, Liu TT, Shih D, Remke M, Schubert S, Bouffet E, Fisher PG, et al. (2014). MRI surrogates for molecular subgroups of medulloblastoma. AJNR Am J Neuroradiol 35, 1263–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DSW, and Parker R (2016). Principles and Properties of Stress Granules. Trends Cell Biol 26, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, et al. (2012). Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 488, 106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnaparkhe M, Wong JKL, Wei P-C, Hlevnjak M, Kolb T, Simovic M, Haag D, Paul Y, Devens F, Northcott P, et al. (2018). Defective DNA damage repair leads to frequent catastrophic genomic events in murine and human tumors. Nature Communications 9, 4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, and Vilo J (2019). g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 47, W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, et al. (2012). Novel mutations target distinct subgroups of medulloblastoma. Nature 488, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychowdhury S, and Chinnaiyan AM (2014). Translating Genomics for Precision Cancer Medicine. Annual Review of Genomics and Human Genetics 15, 395–415. [DOI] [PubMed] [Google Scholar]
- Samir P, Kesavardhana S, Patmore DM, Gingras S, Malireddi RKS, Karki R, Guy CS, Briard B, Place DE, Bhattacharya A, et al. (2019). DDX3X acts as a live-or-die checkpoint in stressed cells by regulating NLRP3 inflammasome. Nature 573, 590–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scala M, Torella A, Severino M, Morana G, Castello R, Accogli A, Verrico A, Vari MS, Cappuccio G, Pinelli M, et al. (2019). Three de novo DDX3X variants associated with distinctive brain developmental abnormalities and brain tumor in intellectually disabled females. Eur J Hum Genet 27, 1254–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinin I, Sie D, Bengtsson H, van de Wiel MA, Olshen AB, van Thuijl HF, van Essen HF, Eijk PP, Rustenburg F, Meijer GA, et al. (2014). DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res 24, 2022–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih J-W, Wang W-T, Tsai T-Y, Kuo C-Y, Li H-K, and Wu Lee Y-H (2012). Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem J 441, 119–129. [DOI] [PubMed] [Google Scholar]
- Snijders Blok L, Madsen E, Juusola J, Gilissen C, Baralle D, Reijnders MRF, Venselaar H, Helsmoortel C, Cho MT, Hoischen A, et al. (2015). Mutations in DDX3X Are a Common Cause of Unexplained Intellectual Disability with Gender-Specific Effects on Wnt Signaling. Am J Hum Genet 97, 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Rifo R, Rubilar PS, Limousin T, de Breyne S, Décimo D, and Ohlmann T (2012). DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J 31, 3745–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Merino D, Nimmervoll B, Gupta K, Wang Y-D, Finkelstein D, Dalton J, Ellison DW, Ma X, Zhang J, et al. (2015). Cross-Species Genomics Identifies TAF12, NFYC, and RAD54L as Choroid Plexus Carcinoma Oncogenes. Cancer Cell 27, 712–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H, Gallouzi IE, Chebli K, Capony JP, Mouaikel J, van der Geer P, and Tazi J (2001). RasGAP-associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation-dependent localization. Mol Cell Biol 21, 7747–7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unamuno X, Gómez-Ambrosi J, Ramírez B, Rodríguez A, Becerril S, Valentí V, Moncada R, Silva C, Salvador J, Frühbeck G, and Catalán V (2019). NLRP3 inflammasome blockade reduces adipose tissue inflammation and extracellular matrix remodeling. Cell Mol Immunol, 10.1038/s41423-41019-40296-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakilian H, Mirzaei M, Sharifi Tabar M, Pooyan P, Habibi Rezaee L, Parker L, Haynes PA, Gourabi H, Baharvand H, and Salekdeh GH (2015). DDX3Y, a Male-Specific Region of Y Chromosome Gene, May Modulate Neuronal Differentiation. J Proteome Res 14, 3474–3483. [DOI] [PubMed] [Google Scholar]
- Valentin-Vega YA, Wang Y-D, Parker M, Patmore DM, Kanagaraj A, Moore J, Rusch M, Finkelstein D, Ellison DW, Gilbertson RJ, et al. (2016). Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Sci Rep 6, 25996–25996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiente-Echeverría F, Hermoso MA, and Soto-Rifo R (2015). RNA helicase DDX3: at the crossroad of viral replication and antiviral immunity. Rev Med Virol 25, 286–299. [DOI] [PubMed] [Google Scholar]
- van de Wiel MA, Kim KI, Vosse SJ, van Wieringen WN, Wilting SM, and Ylstra B (2007). CGHcall: calling aberrations for array CGH tumor profiles. Bioinformatics (Oxford, England) 23, 892–894. [DOI] [PubMed] [Google Scholar]
- Vladoiu MC, El-Hamamy I, Donovan LK, Farooq H, Holgado BL, Sundaravadanam Y, Ramaswamy V, Hendrikse LD, Kumar S, Mack SC, et al. (2019). Childhood cerebellar tumours mirror conserved fetal transcriptional programs. Nature 572, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voet S, Srinivasan S, Lamkanfi M, and van Loo G (2019). Inflammasomes in neuroinflammatory and neurodegenerative diseases. EMBO Mol Med 11, e10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Schwarz SC, Tost J, Trümbach D, Winter P, Busato F, Tacik P, Windhorst AC, Fagny M, Arzberger T, et al. (2018). Epigenome-wide DNA methylation profiling in Progressive Supranuclear Palsy reveals major changes at DLX1. Nature Communications 9, 2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welstead GG, Creyghton MP, Bilodeau S, Cheng AW, Markoulaki S, Young RA, and Jaenisch R (2012). X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proceedings of the National Academy of Sciences of the United States of America 109, 13004–13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore C, Eberhart DE, and Curran T (2000). The Normal patched Allele Is Expressed in Medulloblastomas from Mice with Heterozygous Germ-Line Mutation of patched. Cancer Research 60, 2239–2246. [PubMed] [Google Scholar]
- Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, Schuller U, Machold R, Fishell G, Rowitch DH, et al. (2008). Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell 14, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Kleiner S, Wu J, Sah R, Gupta RK, Banks AS, Cohen P, Khandekar MJ, Boström P, Mepani RJ, et al. (2012). TRPV4 is a regulator of adipose oxidative metabolism, inflammation, and energy homeostasis. Cell 151, 96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Xing L, Rossoll W, Wichterle H, Singer RH, and Bassell GJ (2006). Multiprotein complexes of the survival of motor neuron protein SMN with Gemins traffic to neuronal processes and growth cones of motor neurons. J Neurosci 26, 8622–8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhao B, Yan T, Hao A, Gao Y, Li D, and Sui G (2018). G-quadruplex structures at the promoter of HOXC10 regulate its expression. Biochimica et biophysica acta Gene regulatory mechanisms 1861, 1018–1028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA and DNA sequencing datasets generated during this study have been submitted to the GEO database (Accession ID: GSE147178 for RNA-sequencing and GSE147069 for DNA-sequencing). Other RNA sequencing sets that were previously published are referenced and available as indicated in the respective papers.


