Figure 2. Mechanism of AT1R activation by AngII and biased agonists.
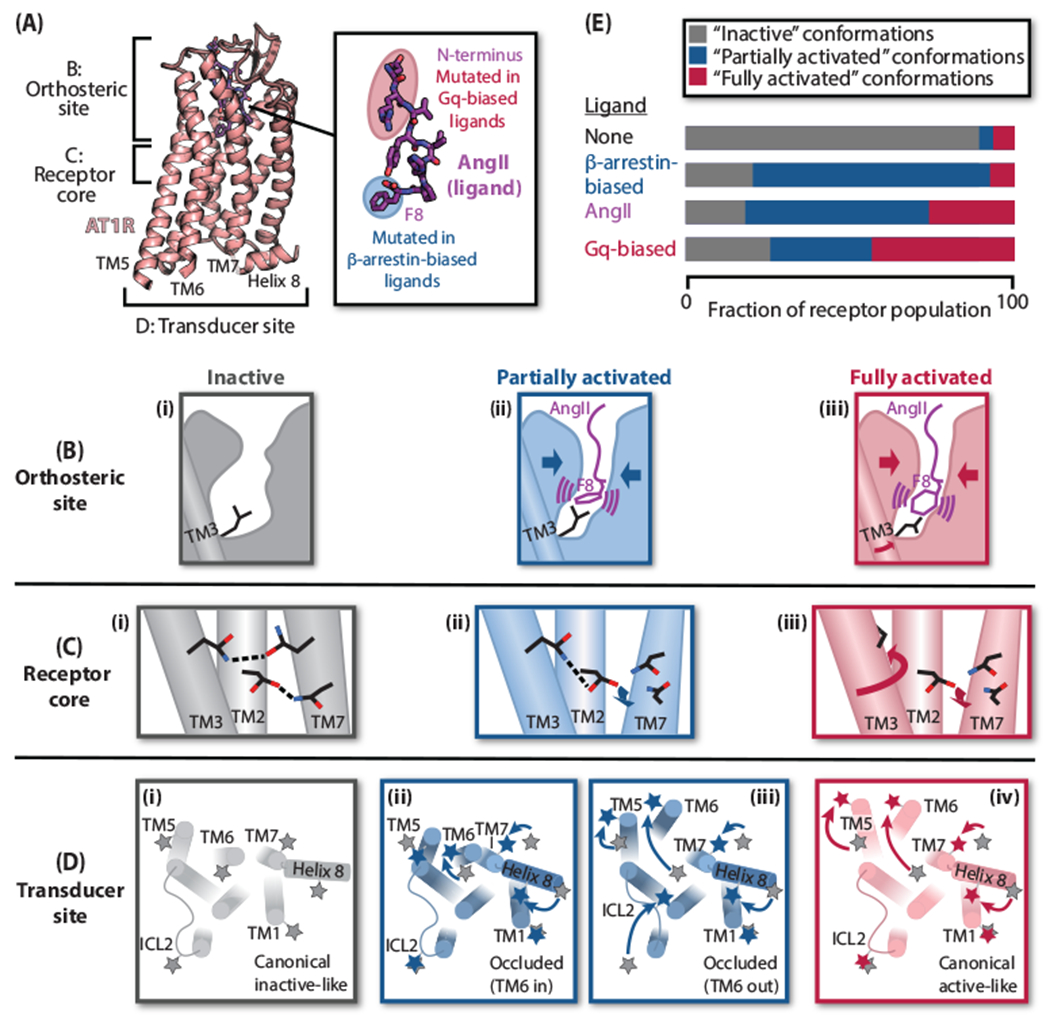
(A) Overview of AT1R-AngII structure (PDB: 6OS0) and structure-activity relationships of biased AngII analogs. (B)-(D) Cartoon representations of the distinct states of the AT1R orthosteric site, receptor core, and transducer site—classified as “inactive” (gray), “partially activated” (blue), and “fully activated” (red)—observed by crystallography, spectroscopy, and MD simulations. (B) Compared to the inactive state (i), binding of either AngII or β-arrestin-biased ligands (lacking F8) causes contraction of the orthosteric site (ii). Certain AngII F8 conformations promote an additional rotation of TM3 (iii). (C) A hydrogen-bonding network involving TMs 2, 3, and 7 in the receptor core stabilizes the inactive state (i). In structures with β-arrestin-biased ligands (ii) and AngII (iii), the movement of TM7 breaks its connection to this network. In the AngII-bound structure, the additional rotation of TM3 removes the key residue from the network (iii). (D) Besides transducer site conformations consistent with the canonical inactive (i) and fully activated (iv) GPCR conformations, DEER spectroscopy delineated two partially activated, “occluded” conformations—one occluded by virtue of an inward TM6 (ii), and the other due to differences in the second intracellular loop (ICL2) and TM5 (iii). Stars represent nitroxide labels; positions of labels from the inactive state (i) are shown in gray in each panel for reference. (E) Model of AT1R biased ligand activation. β-Arrestin-biased ligands stabilize only partially activated conformations; AngII promotes both partially and fully activated conformations; and Gq-biased ligands stabilize fully activated conformations more strongly than AngII.
