Extended Data Fig. 1.
Genome-wide CRISPR screen for essential hepatovirus host factors.
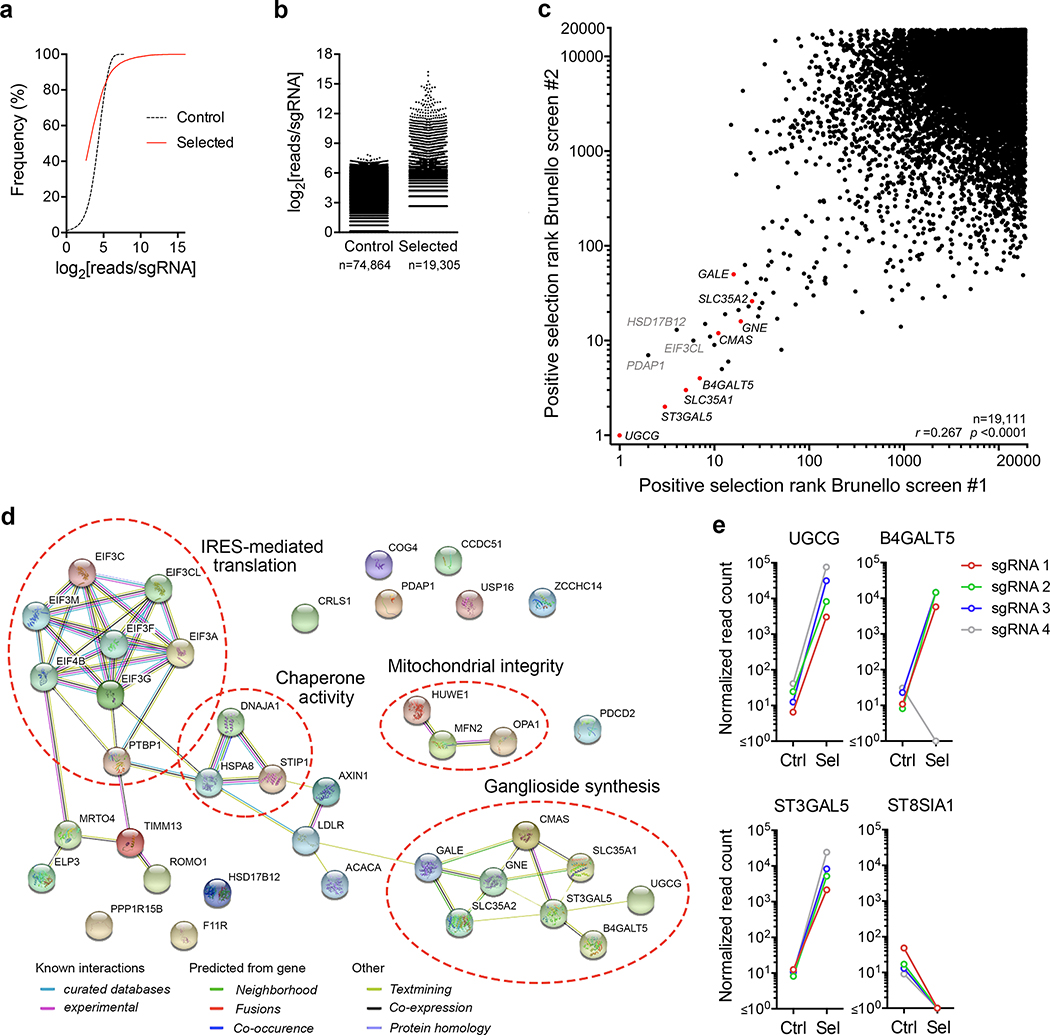
a. Frequency distribution of the number of reads mapping to individual sgRNA integrants in high-throughput sequencing of control (GCV-treated) versus selected (18f-Tat virus-infected and GCV treated) HeLa-tkGFP cells transduced with lentiviruses expressing the Brunello sgRNA library in the first of two independent screens (screen #1). b. Number of reads (log2) mapping to individual sgRNAs in control versus selected cells in screen #1. The total number of individual sgRNAs to which reads mapped is shown at the bottom. c. Scatterplot showing correlation of the positive selection scores accorded genes (n=19,111) on the basis of sgRNA enrichment in two independent screens. Red symbols indicate genes related to NeuNAc or ganglioside synthesis. Spearman’s rank correlation coefficient r=0.267, p < 0.0001. d. STRING analysis of functional associations of proteins encoded by genes comprising the top 39 hits identified in the combined analysis of the two independent screens. Common associations are annotated. Protein-protein interaction enrichment p-value = 3.11 ×10−15. e. Normalized counts of reads mapping to sgRNAs targeting selected genes involved in ganglioside synthesis (see main manuscript Fig. 1c) in control versus selected cell populations.