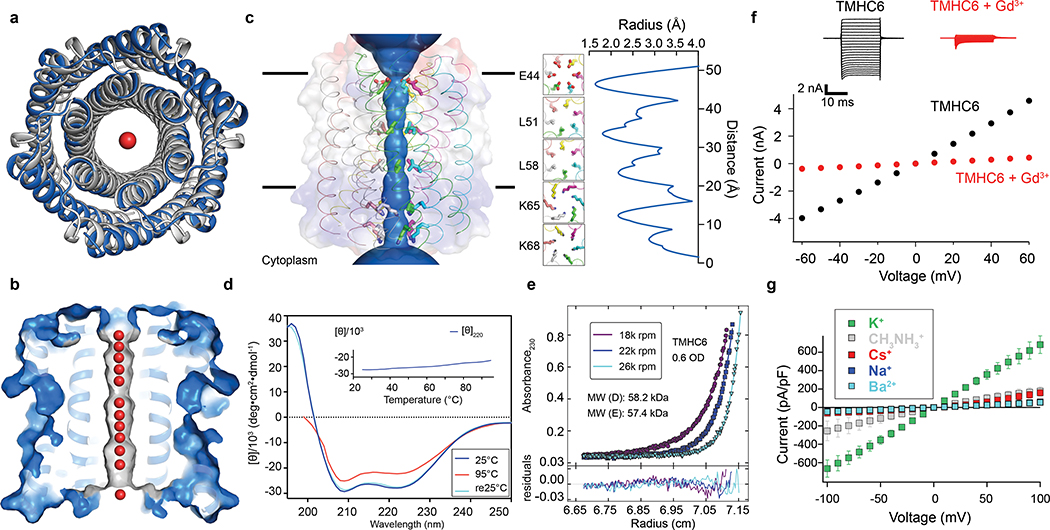
Figure 1. The X-ray crystal structure of the water-soluble hexameric WSHC6 and the ion conductivity of the 12-helix TMHC6 transmembrane channel.
(a) Superposition of backbones of the crystal structure (blue) and the design model (gray) of WSHC6. The C-ɑ RMSD between the crystal structure and the design model is 0.89 Å. (b) The cross section of the WSHC6 channel. A chain of water molecules (red) occupies the central pore (Extended Data Fig. 1e). (c) TMHC6 channel model. The permeation path, calculated by HOLE20, is illustrated by the blue surface in the left panel. Constriction sites along the channel are the E-ring (E44), the K-rings (K65, K68), and two intervening L-rings (L51, L58). Right: the radius of the pore along the permeation path. (d) CD spectra and temperature melt (inset) of the TMHC6 channel. No apparent unfolding transition is observed up to 95°C. Re25°C spectrum is taken when the sample cools back to 25°C after the thermal melt scan. (e) AUC sedimentation-equilibrium curves at three different rotor speeds for TMHC6 yield a measured molecular weight of ~58 kDa consistent with the designed hexamer. “MW (D)” and “MW (E)” indicate the molecular weight of the oligomer design and that calculated from the experiment, respectively. (f) Conductivity in whole-cell patch-clamp experiment on insect cells expressing TMHC6. The channel blocker Gd3+ diminished ion conductance to that of untransfected cells. (g) TMHC6 has considerably higher conductance for K+ than Na+, Cs+, CH3NH3+ and Ba2+. 10 cells were measured for each permeant ion species and the mean (data points) and standard error of measurement (s.e.m., error bars) are plotted.