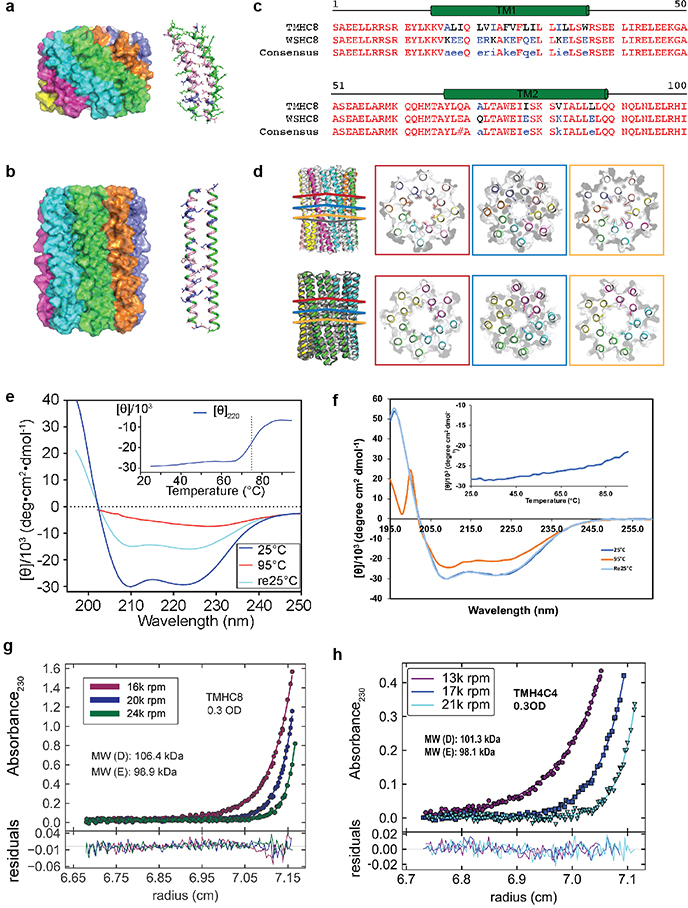
Extended Data Fig. 5 |. Design and additional characterizations of the designed 16-helix pores.
(a & b) Design models from the first and second rounds of water-soluble designs. The monomers of the first round designs (a, 70 amino acids) are considerably smaller than those of the second round (b, 100 amino acids). (c) Sequence alignment of TMHC8 with WSHC8. (d) Pore-lining residues in WSHC8 and TMH4C4. The crystal and cryoEM structures are in colors. The design models are in gray. Top row: The lumen of WSHC8 pore is freely water-accessible, so the residues inside the pore are all polar. Shown in the figure are three representative layers of the pore-lining residues in the crystal structure, Glu69 ring (red), Lys80 ring (blue), and Glu87 (orange). The missing heavy atoms of these flexible residues are built using Rosetta with backbone constraints. Bottom row: three pore-lining layers in the cryo-EM structure of TMH4C4 corresponding to the three layers in the top row. Glu69 and Glu87 are replaced with glutamine and leucine, respectively. (e & f) CD spectra and thermal stability of 16-helix transmembrane pores. An unfolding transition is observed at ~75°C for TMHC8 (e). TMH4C4 (f) is thermally stable up until 95°C. (g & h) Representative AUC sedimentation-equilibrium curves of 16-helix transmembrane nanopores. By fitting the data globally as a single ideal species in solution, TMHC8 is shown to form complexes with a MW of 98.9 kDa, which is in between the MWs of a heptamer and an octamer. The MW of TMH4C4 is determined to be 98.1 kDa, very close to the MW of a tetramer. ‘MW (D)’ refers to the molecular weight of the oligomer design and ‘MW (E)’ refers to the molecular weight determined in the experiment.