Fig. 2: Analysis of p53 function in TP53 G334R mutant cell lines.
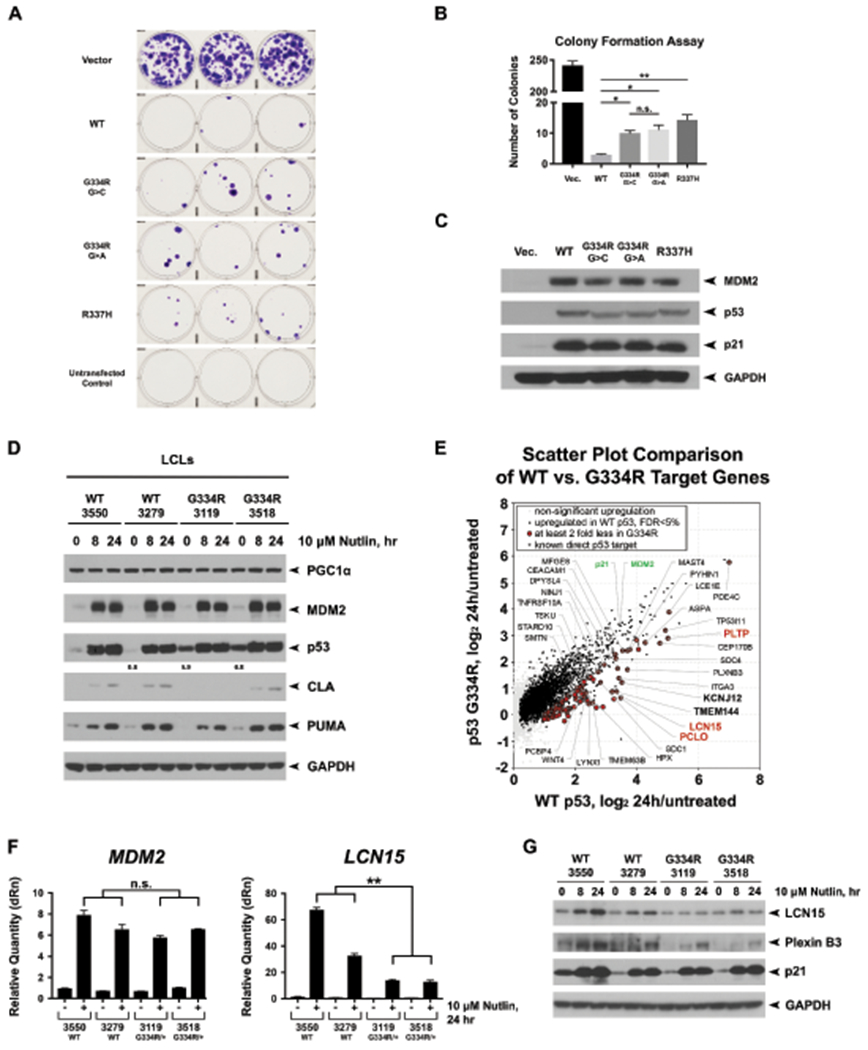
(a) Colony formation assays in H1299 cells transfected with wild-type TP53 (WT), CMV vector alone, and expression constructs containing the c.1000G>C;p.G334R allele (G334R G>C), the c.1000G>A;p.G334R allele (G334R G>A), the p.R337H allele, as well as mock transfected cells. Colonies stained with crystal violet and counted as per inset. Shown are representative images of triplicate wells from the same experiment. All experiments were performed in triplicate and the average numbers of colonies from two biological and three technical replicates are depicted in (b), including standard deviation. (c) Confirmation that transfected H1299 cells expressed equivalent levels of each p53 protein; the slight shift in mobility of p53 is due to Pro72Arg, which does not alter colony suppression. (d) Two TP53 wild-type lymphoblastoid cell lines (LCLs) and LCLs made from the cousin pair of Family G334R-1 were treated for the indicated times with 10μM Nutlin-3a (Nutlin). Western blot of PGC1α, MDM2, p53, cleaved Lamin A (CLA), PUMA, and GAPDH. (e) Scatter plot highlighting the top differentially regulated genes between WT and G334R/+ cells treated with Nutlin. The data depict average changes of two replicates from two WT and p.G334R/+ LCLs treated with 10 μM Nutlin-3a (Nutlin) for 24 hours versus untreated condition. Known direct p53 targets with significantly less response of at least 2 fold in p.G334R/+ cells are highlighted. (f) Quantitative PCR analysis of MDM2 and LCN15 expression levels in two WT and two p.G334R LCLs after 24 hours of 10 μM Nutlin. (g) Western blots of LCN15 and PLXNB3. Data shown are representative of three technical replicates and two biological replicates. *p<0.05. **p<0.01. ***p<0.001
