Fig. 3: Structural analysis of p53-G334R tetramer.
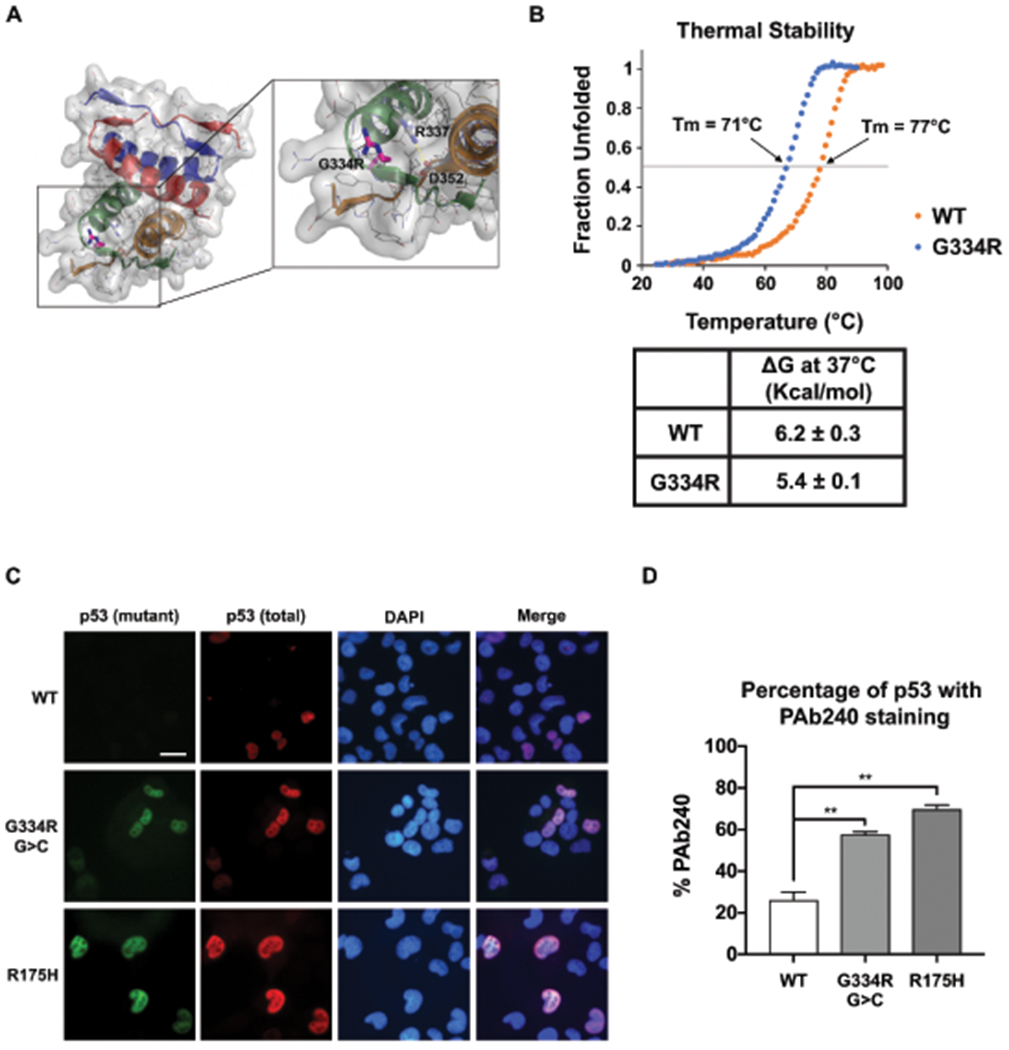
(a) Structural modelling analysis of p53-G334R mutant tetramer. Pymol analysis (PDB, Protein data Bank code 1PES) showing the tetramerization domain comprising residues 310-360. G334 is in a surface exposed loop. Carbon, Nitrogen, and Oxygen atoms are depicted in gray, blue, and red, respectively. The glycine to arginine mutation at position 334 is shown in magenta sticks. The side-chains of R337 and D352 are shown as sticks to highlight the close proximity of this salt bridge to residue 334. The salt bridge is denoted with a dashed yellow line. The surface depicts the space occupied by the non-mutated protein. The cartoon representation of each monomer subunit are colored differently to highlight the symmetry of the tetramer. (b) Thermal stability analysis of wild-type (WT) versus p53-G334R tetramer demonstrating fraction of the tetramer that is denatured with increasing temperature. Inset demonstrates the deltaG of WT versus p53-G334 mutant tetramer. (c) H1299 cells were transfected with TP53 vector (WT) and vectors containing the c.1000G>C;p.G334R allele (G334R G>C), the c.1000G>A;p.G334R allele (G334R G>A), and the p.R337H allele. Cells were subjected to immuno-fluorescence (IF) analysis of total (FL-393) and mutant conformation (pAb240) p53 staining. All experiments were done in triplicate. (d) Quantification of the fraction of cells containing any staining for pAb240 and mutant conformation (pAb240) p53 staining.
