Figure 4 |. PD-L1 interacts with HIP1R to engage Clathrin-dependent endocytosis.
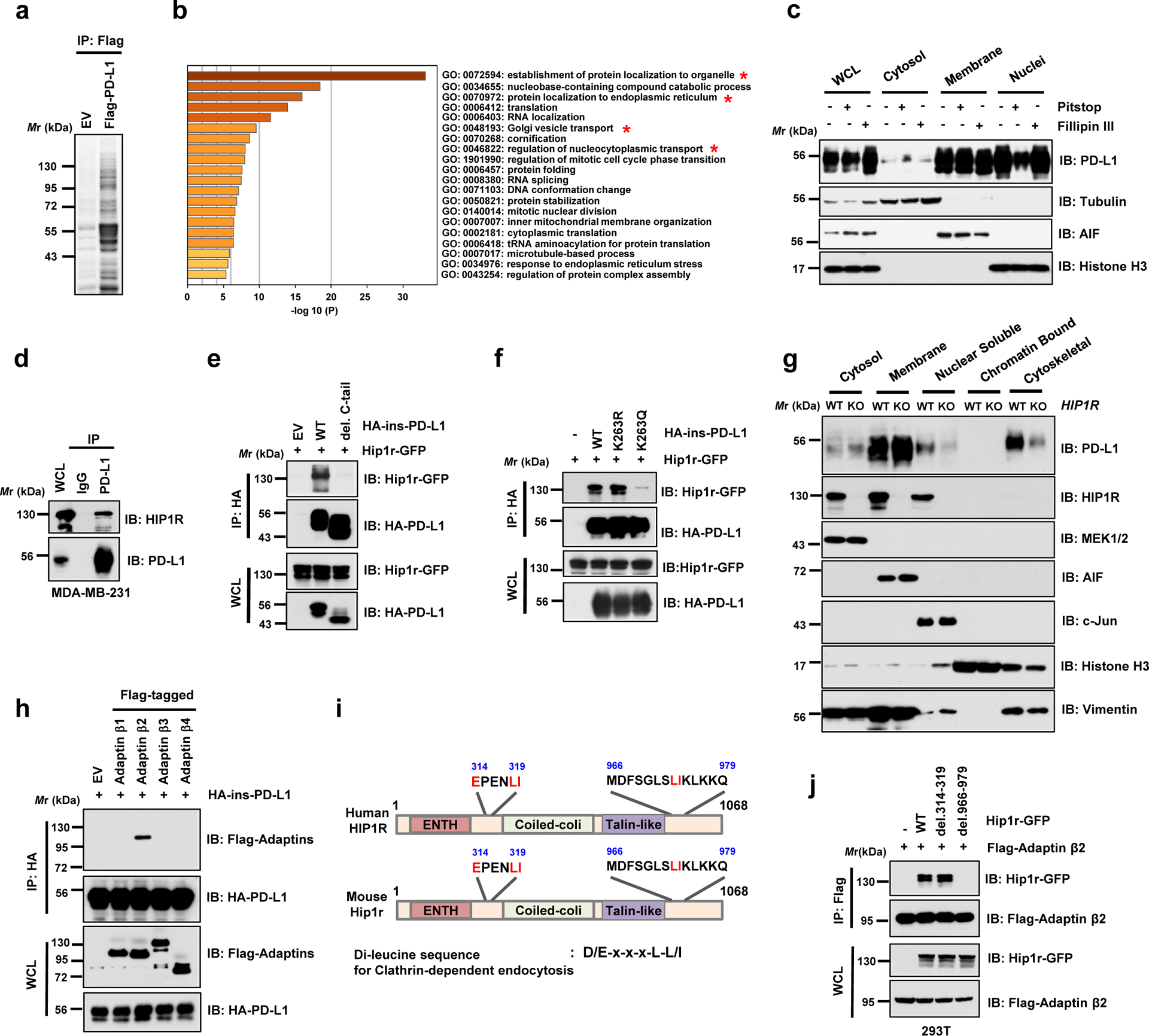
a. Anti-Flag IPs coupled with mass spectrometry analysis to identify PD-L1 interacting proteins in 293T cells, n=2 biologically independent experiments. b. Results from mass spectrometry analysis in a were analyzed for GO term enrichment. Red stars denote pathways associated with protein translocation. n=2 independent experiments with similar results. P values were calculated using hypergeometric test. c. MDA-MB-231 cells were treated with 5 μM Pitstop or 10 μg/ml Fillipin III for 15 min, followed by fractionation analysis. d. IB analysis of WCL and anti-PD-L1 IPs derived from MDA-MB-231 cells. e. IB analysis of WCL and anti-HA IPs derived from 293T cells transfected with mouse Hip1r-GFP and HA-ins-PD-L1 WT or its del. C-tail mutant. f. IB analysis of WCL and anti-HA IPs derived from 293T cells transfected with mouse Hip1r-GFP and HA-ins-PD-L1 WT, K263R or K263Q mutants. g. Fractionation analysis for PD-L1 in MDA-MB-231 WT or HIP1R KO cells. h. IB analysis of WCL and anti-HA IPs derived from 293T cells transfected with HA-ins-PD-L1 and indicated Adaptin constructs. i. Schematic illustration of human and mouse HIP1R protein domains and candidate di-leucine sequences. j. IB of WCL and anti-Flag IPs derived from 293T cells transfected with Flag-tagged Adaptin β2 (AP2B1) and indicated di-leucine sequence deleted constructs.
The western blots in c-h and j were performed for n=2 independent experiments with similar results. Unprocessed immunoblots are shown in Source Data Fig. 4.
