Figure 1. ABP Design and ABPP Workflow.
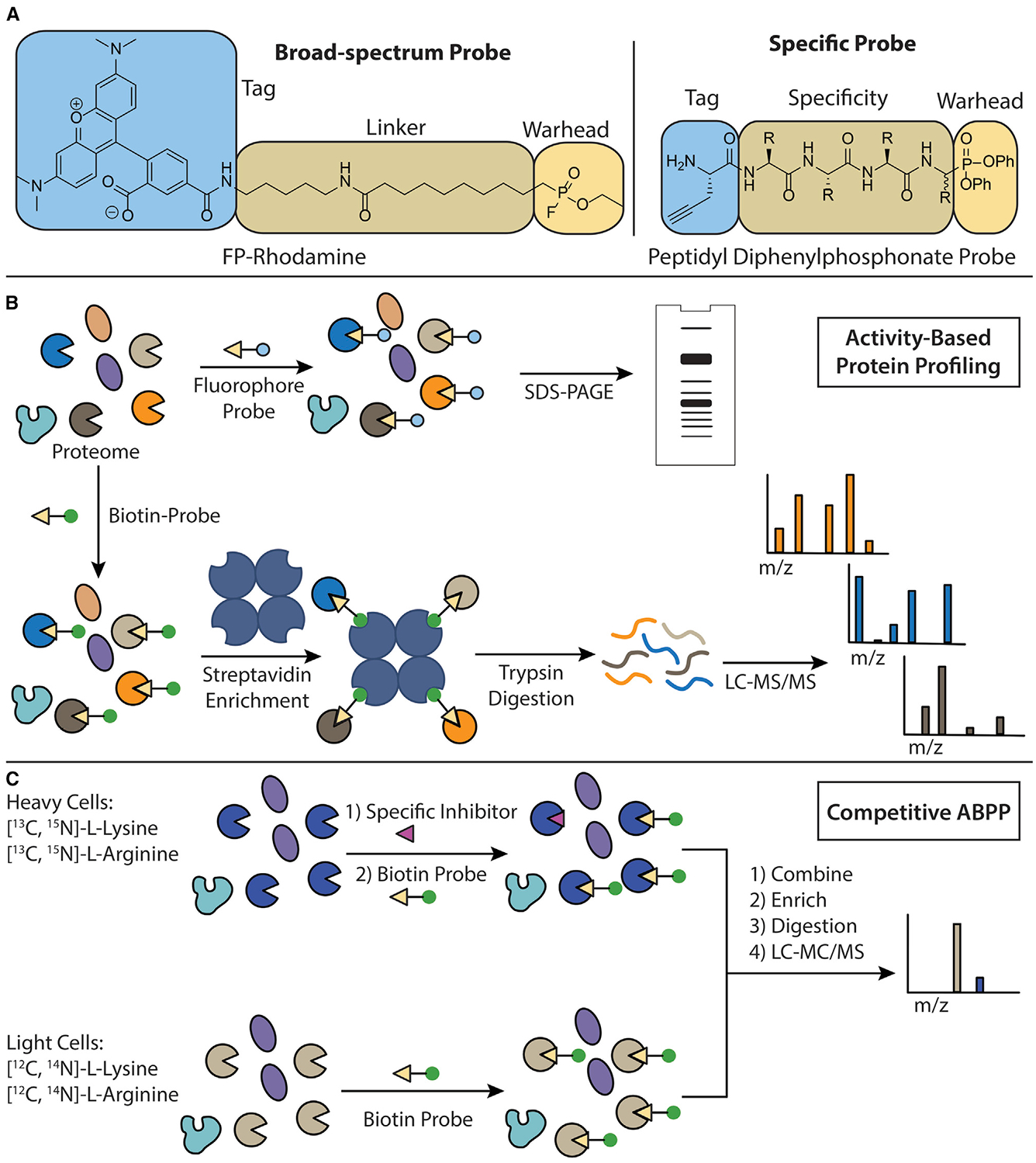
(A) Components of an activity-based probe. The structure of the broad-spectrum FP-rhodamine probe (left). A peptidyl diphenylphosphonate probe scaffold (right). The electrophilic warhead, linker/specificity element, and tag are highlighted.
(B) The workflow of a typical ABPP experiment. Cells or cell lysates are incubated with an ABP. If a fluorescent probe is used, then proteomes can be separated by SDS-PAGE and probe-labeled proteins visualized using in-gel fluorescence. If a biotin-tagged probe is used, then probe-labeled proteins are immobilized on streptavidin resin, digested into tryptic peptides, and subjected to liquid chromatography-tandem mass spectrometry for target identification.
(C) The workflow of a stable isotope competitive ABPP experiment. Cells are grown in the presence of isotopically distinct “heavy” or “light” arginine and lysine. Heavy labeled cells are then pre-treated with an inhibitor before incubation with a broad-spectrum biotinylated ABP. Light labeled cells are pre-treated with a DMSO control before incubation with the same broad-spectrum biotinylated ABP. The heavy and light lysates are mixed together and the same workflow as described in (B) is followed. The heavy-to-light ratio of peptides is then used as a parameter to quantify the selectivity of inhibition of protein targets by the compound of interest.
