Figure 5. Trp2656.48 is part of a string of closely packed aromatic amino acids. See also Figure S5.
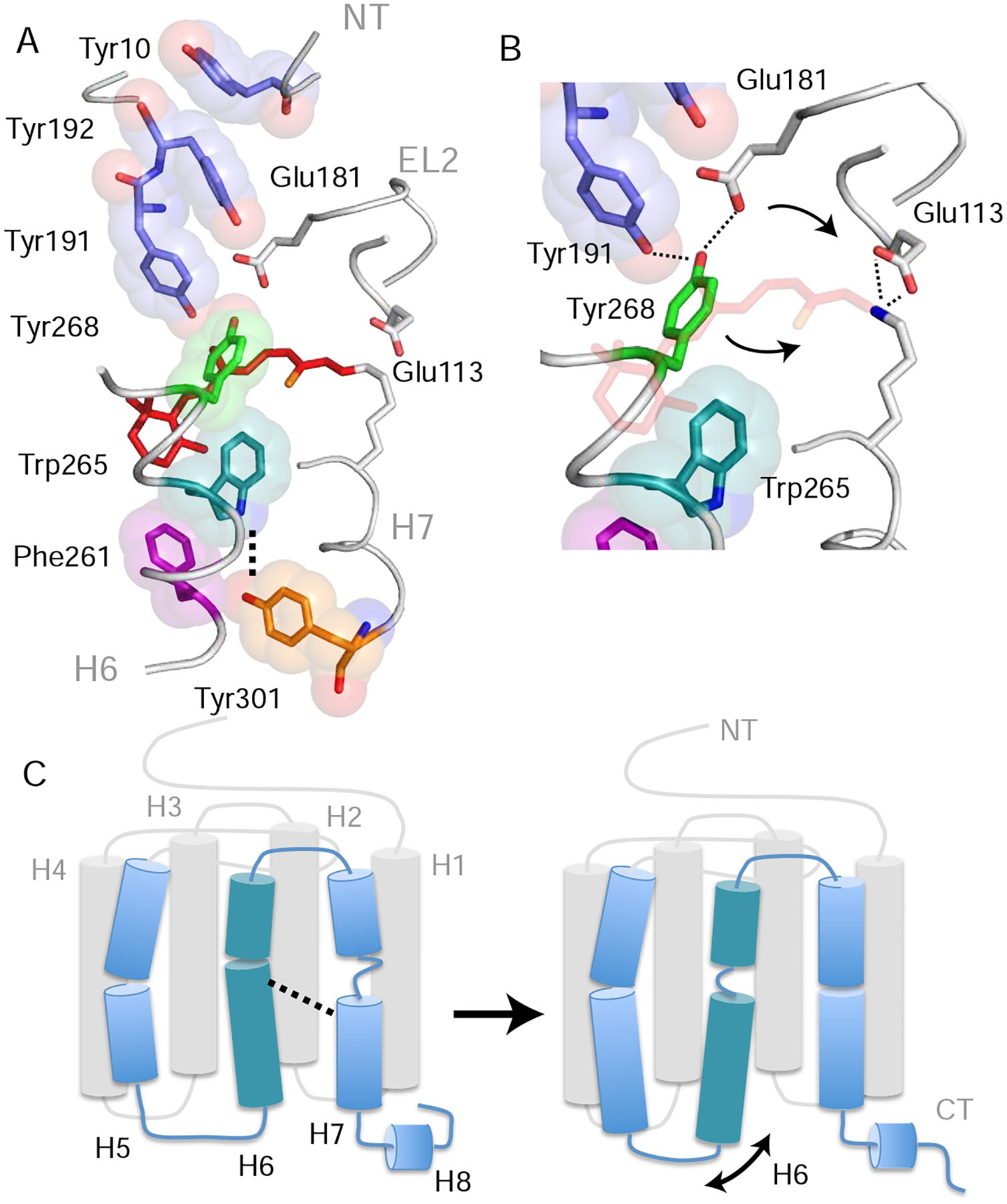
(A) Structure of rhodopsin (PDB: 1GZM) showing the packing of aromatic amino acid side chains stretching from Tyr10NT on the N-terminus to Tyr3017.48 in the TM core (Li et al., 2004). Lys2967.43 forms a positively charged PSB with the retinal that interacts with counter-ions Glu1133.28 on H3 and Glu181EL2 on EL2. Retinal isomerization and PSB deprotonation influence both the hydrogen-bonding network that includes these glutamate residues as well as Tyr191EL2, Tyr192EL2 and Tyr2686.51 (Kimata et al., 2016a). (B) Expansion of panel (A) in the region of Tyr2686.51. Tyr2686.51 is one of the most conserved residues in the visual receptor subfamily. Its position is defined by aromatic packing against Trp2656.48 and hydrogen-bonding interactions with Tyr191 EL2 and Glu181 EL2. Retinal isomerization (and conversion to Meta I) leads to a shift of the counterion from Glu1133.28 to Glu181EL2 (Yan et al., 2003) and movement of Tyr2686.51 toward the positively charged retinal PSB (Kimata et al., 2016a). (C) Summary of the overall structural changes in H6 and H7 described above. Retinal isomerization and PSB deprotonation lead to unraveling of the H6 helix in the region of Trp2656.48 and helix formation of H7 between Phe2947.41 and Tyr3017.48. The loss of helical structure in H6 facilitates an increase in dynamics of the intracellular end of the helix. Models were made using PyMOL 1.8.07 and the figure was assembled using Adobe Illustrator 15.0.0.
