Figure 3. Parallel Reaction Monitoring Assay Development.
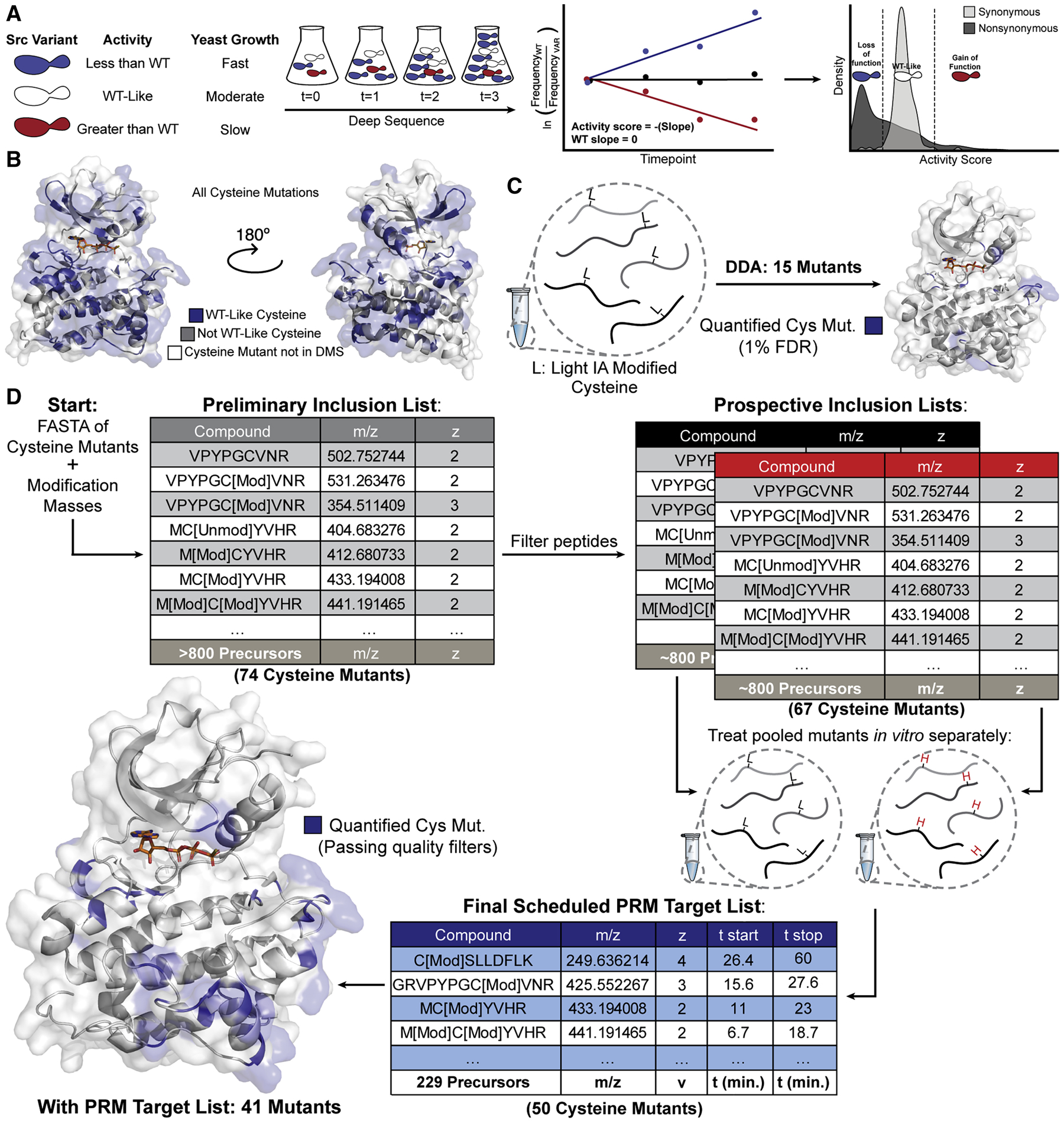
(A) Schematic of the yeast growth-based DMS of Src’s kinase domain (Ahler et al. 2019). (B) Binned activity scores for all Cys substitutions mapped onto Src’s kinase domain (PDB: 3DQW). (C) Initial attempt to identify Cys mutants without enrichment or targeting yielded 15 mutant IDs with mutant peptide FDR<1%. (D) Schematic of PRM assay development. Candidate precursors were identified for all mutant Cys residues by inputting mutant FASTA sequences and modification masses of heavy and light IA into Skyline. Precursors were filtered by removing potentially problematic peptides resulting in prospective inclusion lists for each mass label . Each prospective DDA inclusion list was run in triplicate and resulting spectra were aggregated using Skyline to generate a spectral library and a corresponding Scheduled PRM Target List.
