Figure 6. Incomplete GFHK99 virus genomes are present but not sufficiently abundant to account for observed reassortment in MDCK cells.
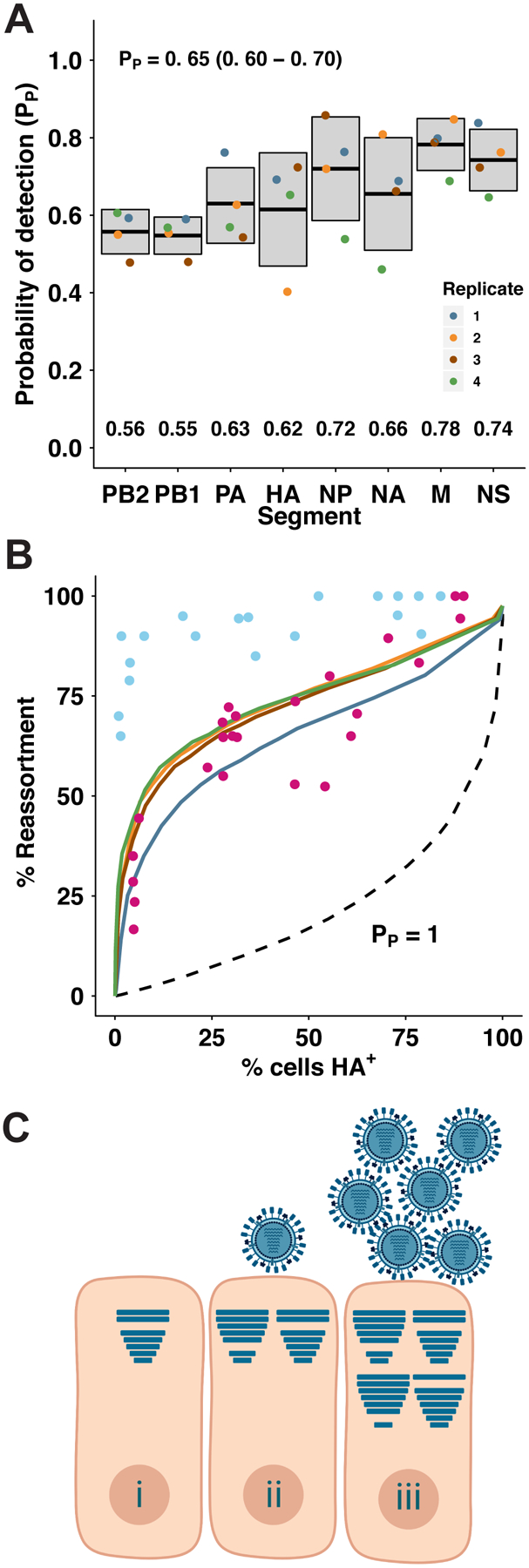
Incomplete viral genomes were quantified experimentally by a single-cell assay which relies on the amplification of incomplete viral genomes of GFHK99 WT virus (0.018 PFU per cell) by a genetically similar coinfecting virus, GFHK99 VAR2. Based on the rate of detection of GFHK99 WT virus segments in this assay, the probability that a given segment would be present and replicated in a singly infected MDCK cell is reported as PP. A) Summary of experimental PP data. n = 4 biological replicates, distinguished by color. Horizontal bars and shading represent mean ± standard deviation. Mean PP,i values for each segment are indicated at the bottom of the plot area. Average PP for each experiment was calculated as the geometric mean of the eight PP,i values, and the mean ± standard deviation of those four summary PP values is shown at the top of the plot area. B) Experimentally obtained PP,i values in (A) were used to parameterize a computational model7. Levels of reassortment predicted using the experimentally determined parameters are shown with colors corresponding to points in (A). Levels of reassortment predicted if PP=1.0 are shown with the dashed line. Observed reassortment of GFHK99 WT and VAR viruses in MDCK cells are shown with blue circles. Observed reassortment of GFHK99 WT and VAR viruses in DF-1 cells are shown with pink circles. Observed data are the same as those plotted in Figure 1. C) Model: complementation of incomplete viral genomes is necessary but not sufficient for robust progeny production from mammalian cells infected with GFHK99 virus. Cells infected with single virions often replicate incomplete viral genomes and therefore do not produce viral progeny (i). In mammalian cells, complementation of incomplete viral genomes through coinfection may allow progeny production (ii), but robust viral yields require the delivery of additional genomes to the cell, beyond the levels needed for complementation (iii). Horizontal bars within cells represent segments successfully delivered and replicated.
