Figure 1: Genomic signatures of adaptation.
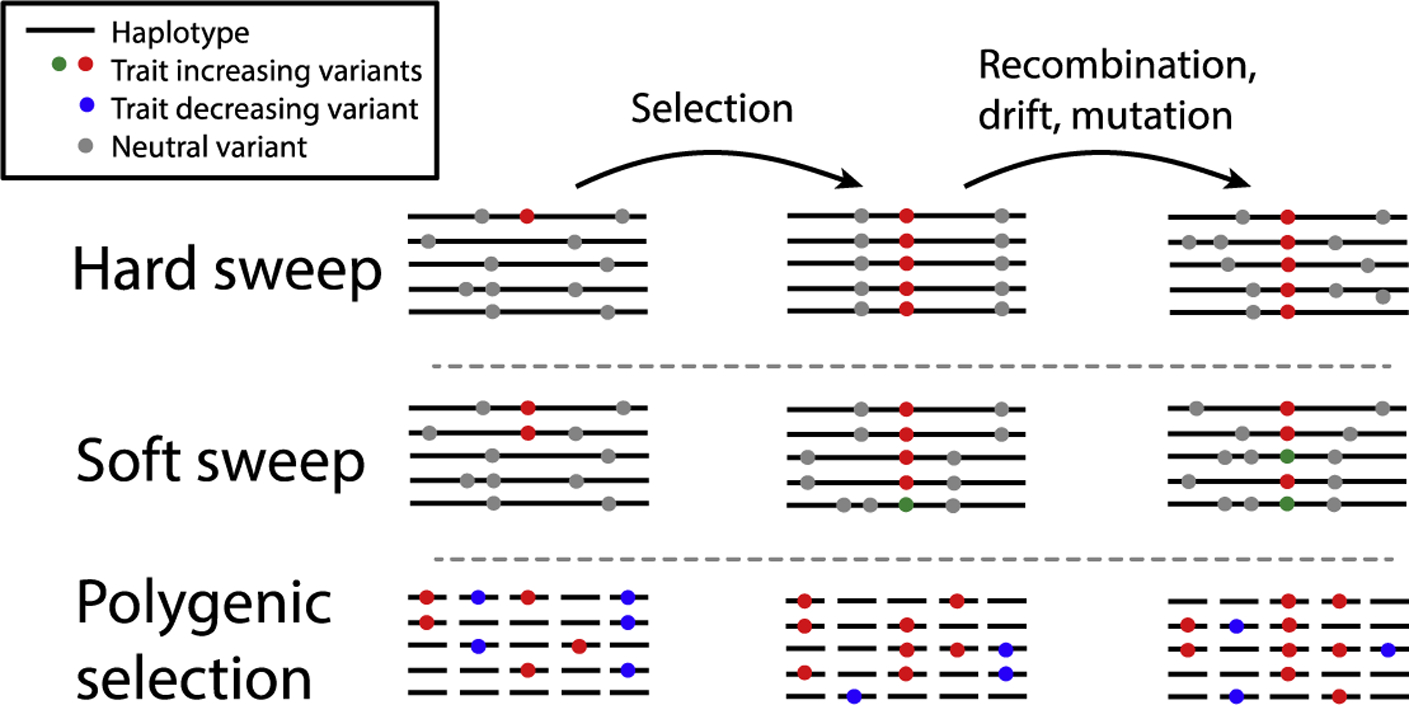
Selection on variants affecting a beneficial trait. In a hard sweep, a new (or rare) mutation (red) on a single haplotype increases rapidly in frequency. Variants on the same haplotype (grey) also increase (“hitchhike”), reducing diversity around the selected site (97). Over time, recombination, drift and mutation break down the sweep signature. In a soft sweep (98), the selected variant may already be present on multiple haplotypes (red), or there may be new selected mutations (green) as the sweep is in progress. Diversity is reduced around the sweep but not by as much as a hard sweep. In practice, this signature may be difficult to distinguish from an incomplete hard sweep. In polygenic adaptation (42), variants at many loci genome-wide change frequency; trait-increasing alleles (red) increase in frequency while trait-decreasing alleles (blue) decrease. This process is essentially a large number of weak soft sweeps, but the effects are too small to be detected at any one locus. Variants drift after selection, but mean shifts in frequency are maintained.
