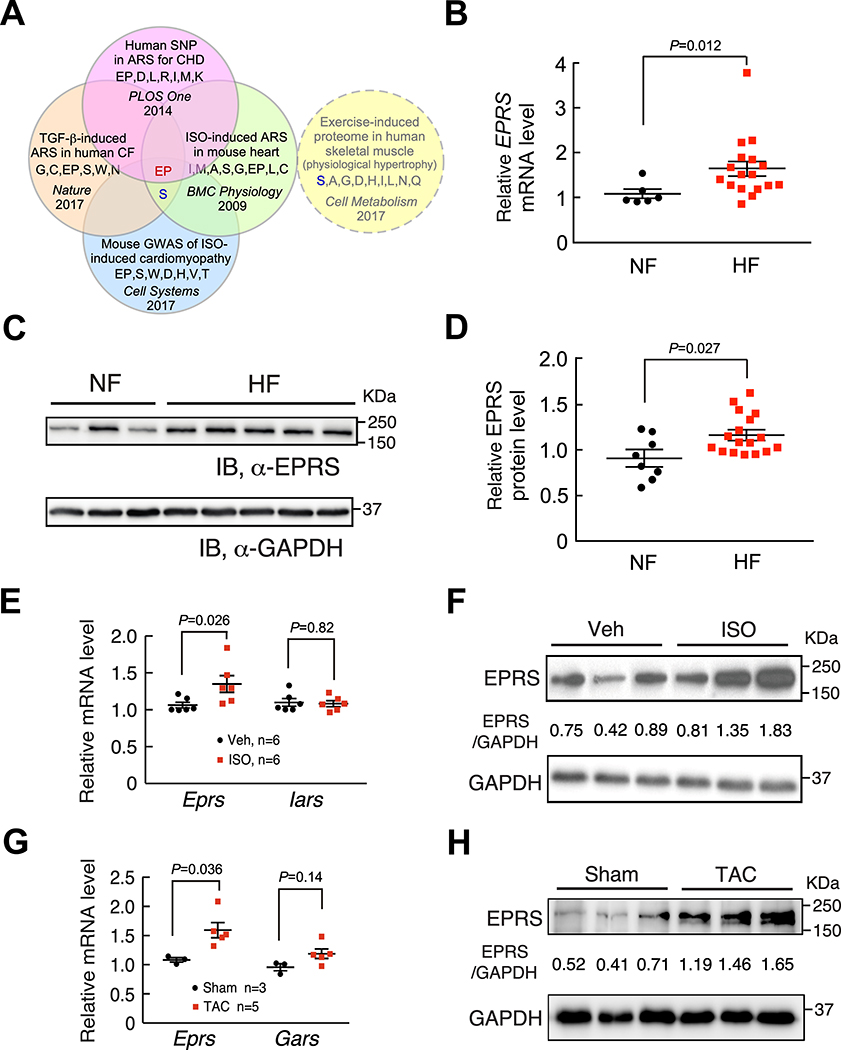
Figure 1. EPRS is upregulated in human and mouse heart failure.
(A) Glutamyl-prolyl-tRNA synthetase (EPRS) is a common hit of upregulated ARSs and ARSs with a genetic association in human and mouse heart disease, but not induced in human physiologic muscle hypertrophy.
(B) EPRS mRNA is increased in failing human hearts (n=17) compared to non-failing donor hearts (n=6). 18S rRNA is used as a loading control.
(C) EPRS protein is increased in failing human hearts compared to non-failing donor hearts. Representative Western blot results are shown.
(D) Quantification of Western blot results from all human heart samples indicates elevated expression of EPRS in failing human hearts (n=17) compared to non-failing donor hearts (n=8). GAPDH is used as a loading control for quantification.
(E-F) EPRS mRNA and protein expression are increased in the hearts from a 4-week ISO infusion induced mouse HF model compared to vehicle infused hearts while Iars (Isoleucyl-tRNA synthetase) mRNA remains unchanged. 18S rRNA and GAPDH are used as loading controls for mRNA and protein quantification, respectively.
(G-H) EPRS mRNA and protein expression are increased in the hearts from an 8-week TAC surgery-induced mouse HF model. Gars (glycyl-tRNA synthetase) is a negative control ARS in the TAC model. 18S rRNA and GAPDH are used as loading controls for mRNA and protein quantification, respectively.
Comparisons were performed by non-parametric unpaired Mann-Whitney test for B, D, E, G.