Figure 2.
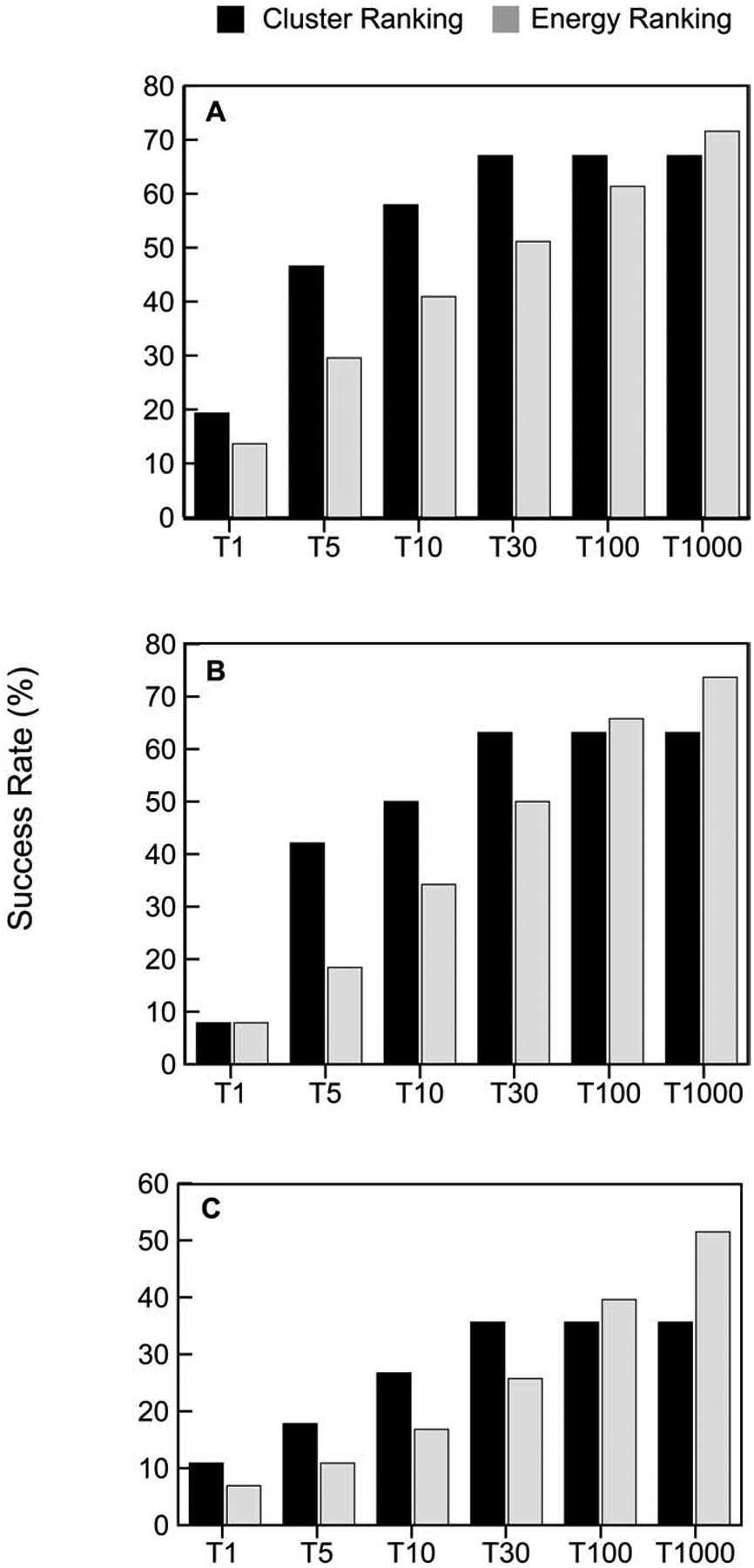
Cluster-based versus energy-based ranking of models. A. Percentage of “good” models in the top 1, top 5, top 10, top 30, and top 1000 predictions for enzyme containing complexes, based on either cluster size or energy value. We consider only up to 30 clusters, and hence the performance of cluster-based selection does not improve beyond 30. Note that energy-based selection of the top 1000 models means that all structures generated by ClusPro are retained. B. Same as A, but for antibody-antigen complexes. C. Same as A, but for “other” type complexes.
