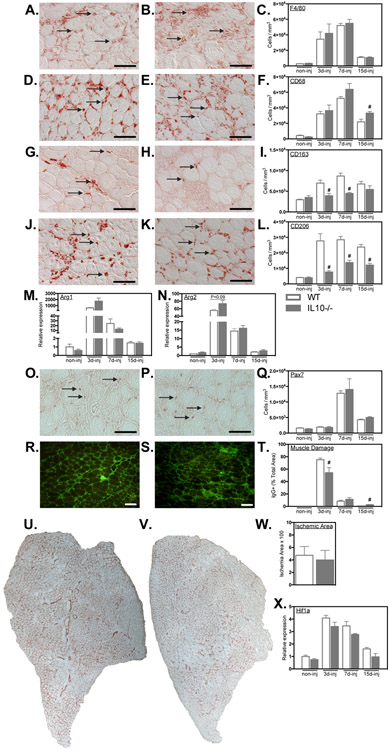
Figure 9.
Il10 mutation attenuated the expansion of M2-biased macrophages in injured muscle affecting muscle injury and repair. A-L: Cross-sections of WT and Il10−/− TA muscles were immunolabeled with anti- F4/80 (A, B), anti-CD68 (D, E), anti-CD163 (G, H) and anti-CD206 (J, K). Representative images from WT (A, D, G, J) and mutant (B, E, H, K) muscles at either 7-dpi. Arrows indicate examples of antibody-labeled leukocytes. Bars = 50 μm. Numbers of F4/80+ (C), CD68+ (F), CD163+ (I) or CD206+ (L) cells per volume of sectioned tissue at 3-, 7- and 15-dpi are summarized in the histograms. M, N: QPCR data showed no change in the induction of Arg1 and Arg2 expression in WT and Il10−/− TA muscles after injury. O, P: Representative images of muscle cross-sections from WT (O) and Il10 mutant (P) muscles at 7-dpi immunolabeled with anti-Pax7. Arrows indicate examples of Pax7+ cells. Bars = 50 μm. Q: Numbers of Pax7+ cells per volume of sectioned tissue at 3-, 7- and 15-dpi. R, S: Representative cross-sections of TA muscles were immunolabeled with anti-mouse IgG (green) to assay for sarcolemma damage at 15-dpi in WT and Il10 mutant muscles. Bars = 100 μm. T: The volume fractions of TA muscles occupied by IgG+ fibers were quantified at 3-, 7- and 15-dpi. # indicates significant difference (P < 0.05) from WT within a time point. P-values based on two-tailed t-test. N = 4-6 for each data set. U, V: Representative images of whole TA muscle cross-sections at 7-dpi immunolabeled for CD31 in WT (U) and Il10 mutants (V). W: Quantification of the volume fraction of the muscle sparsely occupied by CD31+ cells at 7-dpi. X: QPCR analysis of Hif1a transcript expression in WT and Il10 mutant muscles at 3-, 7-, and 15-dpi. # indicates significant difference (P < 0.05) from WT within a time point. P-values based on two-tailed t-test. N = 4-5 for each data set. Data are presented as mean ± SEM.