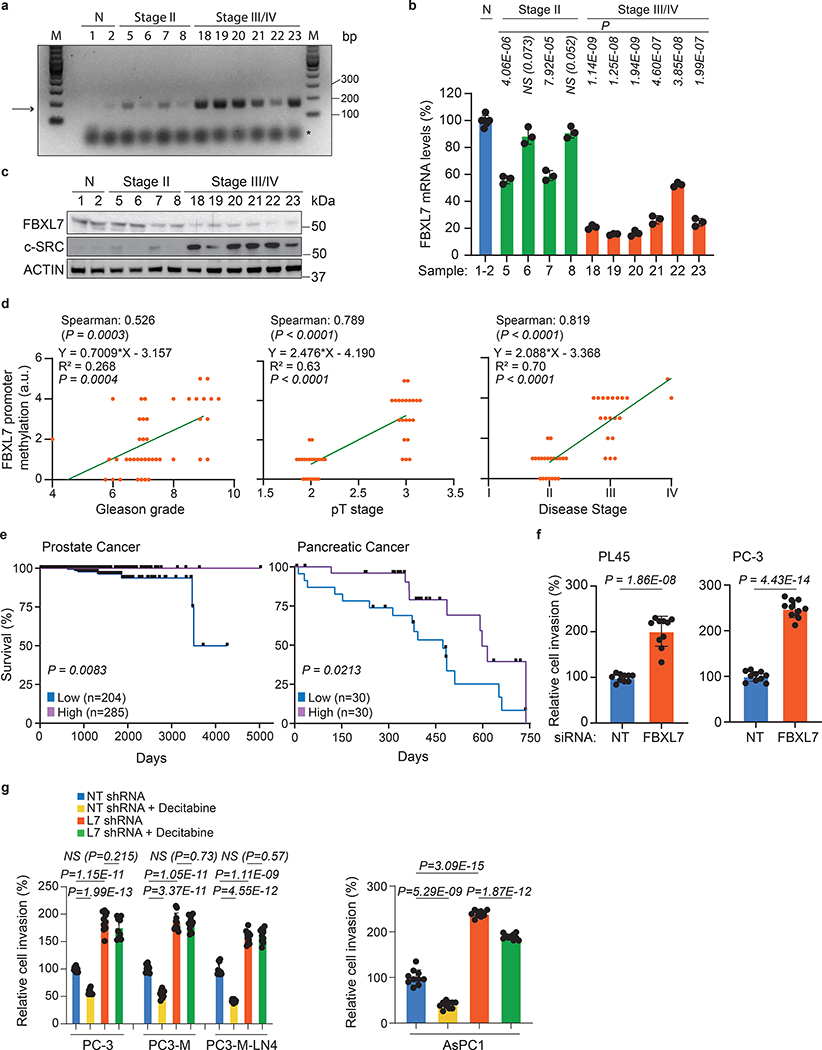
Fig. 2. Low FBXL7 mRNA levels predict poor survival in pancreatic and prostate cancer patients.
a-c, Normal (N) and tumor prostate specimens were analyzed for methylation of the FBXL7 gene promoter (a), FBXL7 mRNA levels (b), and levels of the indicated proteins (c). The arrow points to the methylated FBXL7 promoter, whereas the asterisk indicates the primers’ signal. Details on patients are reported in Extended Data Table 1. n = 12 biologically independent samples. a, c: Two independent experiments were performed, with similar results. b, Mean ± s.d. is shown; n = 3 independent experiments; P values are from unpaired, two-tailed t-test.
d, Correlation analysis of FBXL7 promoter methylation with Gleason grade, pT stage and disease stage for the specimens reported in Extended Data Table 1. n = 41. Two-tailed non-parametric Spearman correlation wad used. The linear regression line is shown.
e, Kaplan-Meier plot showing reduced survival probability of prostate cancer patients with low FBXL7 mRNA levels (n = 204) compared with high (n = 285) FBXL7 mRNA levels (TCGA set), and pancreatic cancer patients with low FBXL7 mRNA levels (n = 30) compared to high (n = 30) FBXL7 mRNA levels (https://www.proteinatlas.org/ENSG0000183580-FBXL7). Log rank (Mantel-Cox) statistical test was used.
f, PL45 and PC-3 cells were transfected with the indicated siRNAs and, after 24 h, re-plated on Matrigel-coated transwells. After 48 h, cells that invaded through Matrigel and migrated on the bottom of the transwells were stained and counted in 10 different fields/well. Each experiment was performed in triplicate. The graph shows quantification from a representative experiment.
g, The indicated cells stably transfected with a pool of shRNAs to FBXL7 or a non-targeting (NT) shRNAs were plated on Matrigel-coated transwells in the presence or absence of decitabine. Each experiment was performed in triplicate. After 48–72 h, cells that invaded through Matrigel were counted as in 2f. The graph shows quantification from a representative experiment. In f, g: two independent experiments were performed with similar results. Mean ± s.d. is shown; n = 10; P values are from unpaired, two-tailed t-test.