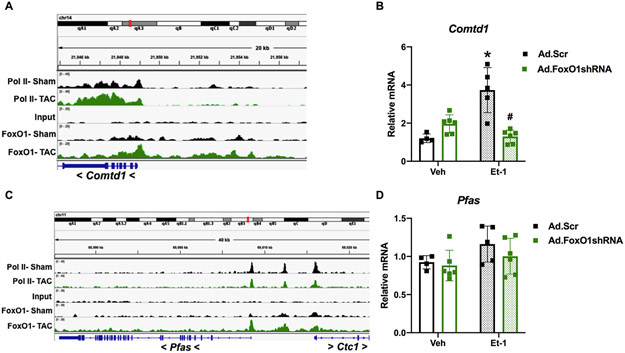
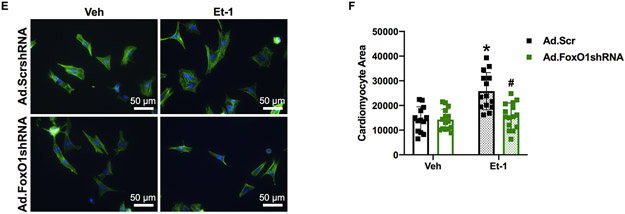
Figure 5. FoxO1 knockdown prevents hypertrophic gene expression and cardiomyocyte hypertophy in vitro.
A-D Forkhead box protein (Fox)O1 and RNA polymerase (pol)II ChIP-Seq were performed in Sham and transverse aortic constriction (TAC) hearts as described in Figure 1. A and C Integrated Genomics Viewer (IGV) software was used to view the fragment densities of pol II and FoxO1 (y-axis) aligned along the gene coordinates (x-axis). Shown are exemplary images for the indicated genes. B and D-F NRVM were treated with vehicle (Veh) or 100 nM endothelin (Et)-1 and/or Ad.FoxO1shRNA or scrambled (Scr) control at an MOI of 10 for 48 h. B and D Total RNA was extracted and qPCR was performed for the indicated genes. The results were plotted. n=4, 5, 6, 6 (Veh, Et-1, Ad.FoxO1shRNA, Ad.FoxO1shRNA+Et-1). E-F Cells were fixed and stained with 488 phalloidin (FITC) and DAPI. Representative images are shown in E. Cell area was quantified using ImageJ software and plotted (F). Cardiomyocytes imaged=13, 14, 15, 15 (Veh/Ad.Scr, Veh/Ad.FoxO1shRNA, Et-1/Ad.Scr, Et-1/Ad.FoxO1shRNA). Error bars represent SEM. *p<0.05 versus Veh/Ad.Scr and #p<0.05 versus Et-1/Ad.Scr using 2-way ANOVA. Ad=adenovirus; shRNA=Short hairpin RNA; Comtd1=Catechol-O-methyltransferase domain containing 1; Pfas=Phosphoribosylformylglycinamidine synthase.