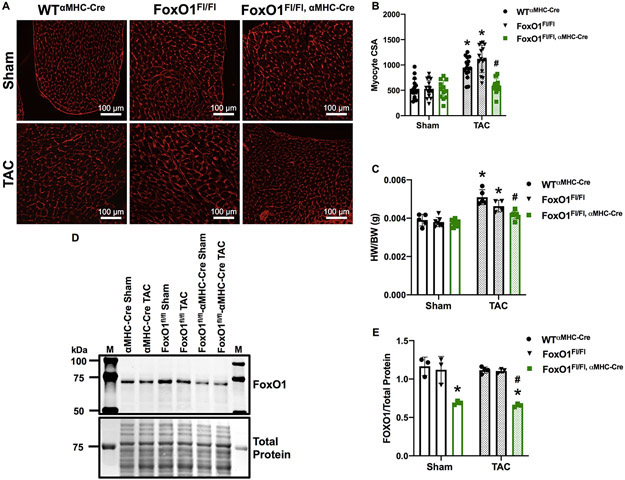
Figure 7. FoxO1 deletion prevents cardiomyocyte hypertophy in vivo.
A-E Adult (12-week-old), male WTαMHC-Cre, FoxO1FL/FL, and FoxO1FL/FL,αMHC-Cre mice were subjected to transverse aortic constriction (TAC) or sham surgery for 7 days. A-B Hearts were harvested, fixed, and stained with 594 WGA. Representative images are shown in A. Cell cross-sectional area (CSA) was quantified using ImageJ software and plotted (B). Cardiomyocytes imaged= 16, 16, 12, 16, 15, 16 (WTαMHC-Cre-Sham, FoxO1FL/FL-Sham, FoxO1FL/FL,αMHC-Cre-Sham, WTαMHC-Cre-TAC, FoxO1FL/FL-TAC, and FoxO1FL/FL,αMHC-Cre-TAC). C Hearts were harvested and weighed. Heart weights (HW) were normalized to body weights (BW) and plotted as a ratio (HW/BW) in grams. n=5, 7, 9, 5, 4, 7 (WTαMHC-Cre-Sham, FoxO1FL/FL-Sham, FoxO1FL/FL,αMHC-Cre-Sham, WTαMHC-Cre-TAC, FoxO1FL/FL-TAC, and FoxO1FL/FL,αMHC-Cre-TAC). D-E Hearts were harvested, lysed, and subjected to Western blotting using the indicated antibody. Signals for forkhead box protein (Fox)O1 were quantified using densitometry, normalized to total protein, and plotted. n=3, 3, 3, 4, 3, 3 (WTαMHC-Cre-Sham, FoxO1FL/FL-Sham, FoxO1FL/FL,αMHC-Cre-Sham, WTαMHC-Cre-TAC, FoxO1FL/FL-TAC, and FoxO1FL/FL,αMHC-Cre-TAC). Error bars represent SEM. *p<0.05 versus WTαMHC-Cre-Sham and #p<0.05 versus WTαMHC-Cre-TAC using 2-way ANOVA. M=marker; WT=Wild type; αMHC=Alpha myosin heavy chain.