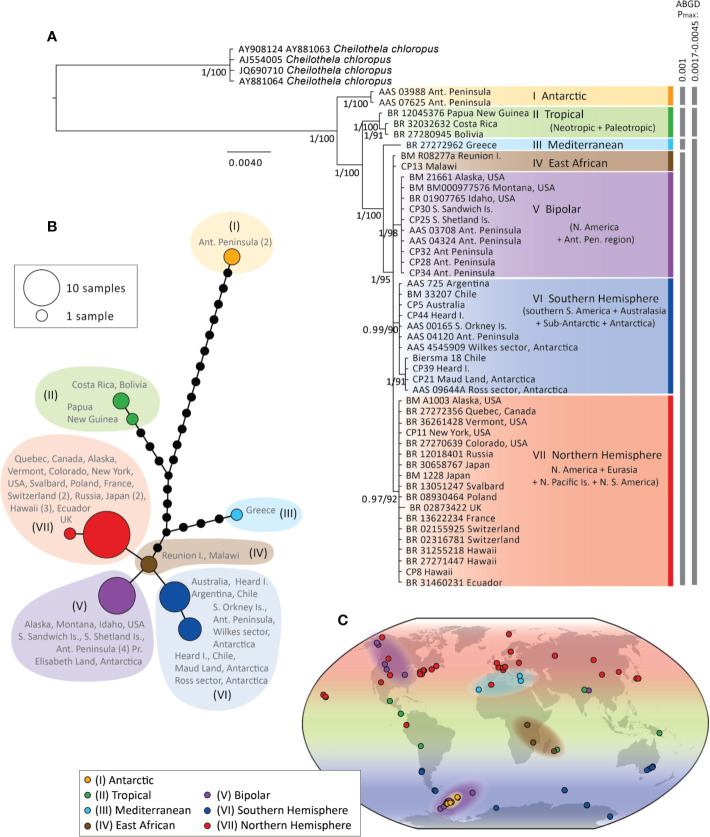
Figure 2.
Bayesian phylogeny (A) and haplotype network (B) for Ceratodon purpureus constructed with a concatenated cpDNA data set (atpB-rbcL+rps4+trnL-F). Posterior probabilities and bootstrap support are shown next to branches (A). The scale bar represents the mean number of nucleotide substitutions per site. Biogeographic clade descriptions (I–VII) and ABGD species-clusters with different Pmax-values are shown next to (A). In (B) haplotype circle sizes and colors correspond to the number of specimens and clades I–VII, respectively. Branches represent mutations between haplotypes, with mutations shown as 1-step edges. (C) represents the sample locations and biogeographic regions of samples in the different clades (I–VII) as interpreted from the concatenated cpDNA data set (A), as well as the placement of samples in phylogenies of single cpDNA markers (e.g. when samples were only represented by one or two single cpDNA marker(s); see also Supplementary Figures S1A–D). For example, while the Mediterranean clade (III) includes just one sample (from Greece) in the concatenated cpDNA data set (A), clade III on the map in (C) includes one more sample from Greece (AY881059) and one from the Canary Islands (BM 27), based on the well-resolved placement of these samples in clade III in the Bayesian phylogenies of atpB-rbcL (Figure S1A) for the former, and rps4 and trnL-F (Figures S1C, D) for the latter, respectively.