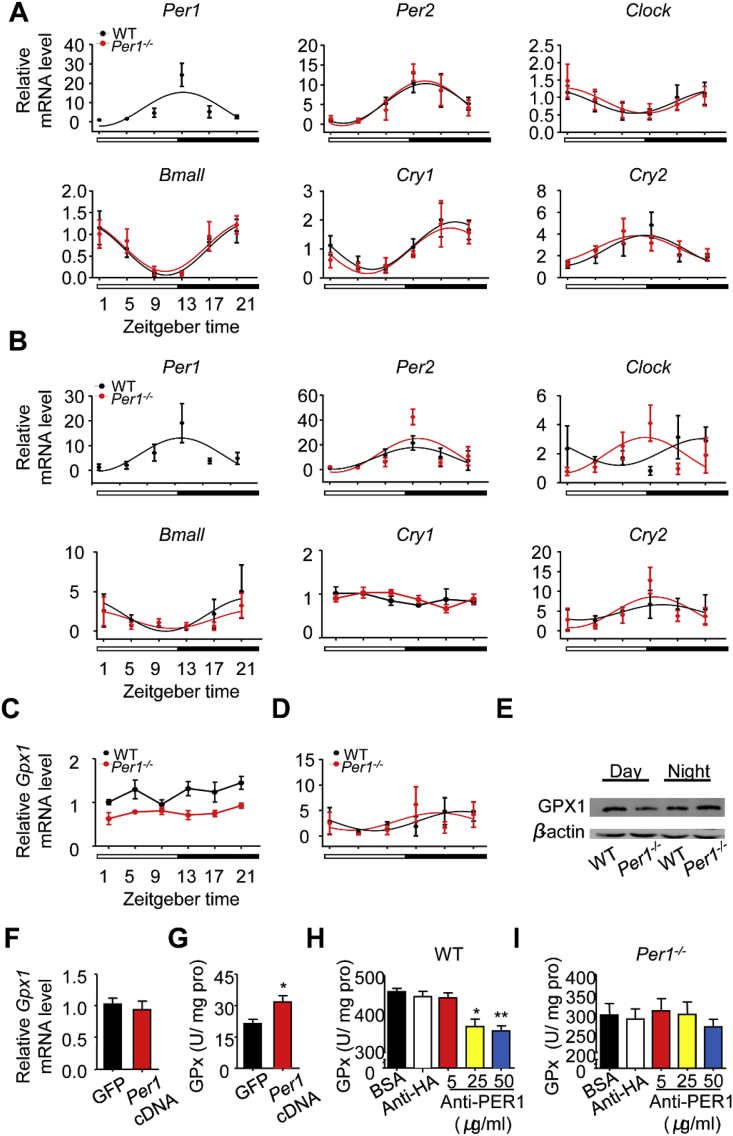
Fig. 4.
PER1 Enhances Gpx Activity through Non-transcriptional Regulation. A, B) The Expression Profiles of Clock Genes in (A) livers and (B) intestines between WT and Per1−/− mice. Quantitative RT-PCR were performed for analyzing mRNA level in WT (black) and Per1−/− (red) mice at indicated times (n = 4/time point/genotype). The white and black bar represents day and night, respectively. No significant differences in these genes' expressions were observed between genotypes. All data were analyzed in detailed Table S4. C, D) Real-time PCR analysis of diurnal mRNA expression of Gpx1 in WT and Per1−/− livers (C) and intestines (D) (n = 4/time point/genotype). The white and black bar represents day and night, respectively. E) Western blot analysis of GPX1 protein level in the livers. Samples were respectively collected at ZT1 and ZT13 between genotypes and pooled samples (n = 3/time point/genotype) were used for each time point. β-actin was used as internal control. F) The mRNA levels of Gpx1 in NIH-3T3 cell line at 48 h after cell were transfected with Per1 cDNA (n = 6 per group). G) The GPx activity in the NIH-3T3 cell line at 48 h after the cells were transfected with Per1 cDNA (n = 6, *p < 0.05, Per1 cDNA versus GFP group). H, I) The GPx activities were measured in fresh liver extracts from WT (H) and Per1−/− (I) mice when various concentrations of PER1 antibody were present. HA antibody (50 μg/ml) as a negative control (n = 4 per group, *p < 0.05, **p < 0.01, compared with control group). Throughout, all data were expressed as the mean ± SEM and male mice for all experiments were maintained on standard chow. Analyses were performed using two-tailed Student's t-test for F, G and one-way ANOVA for H, I. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)