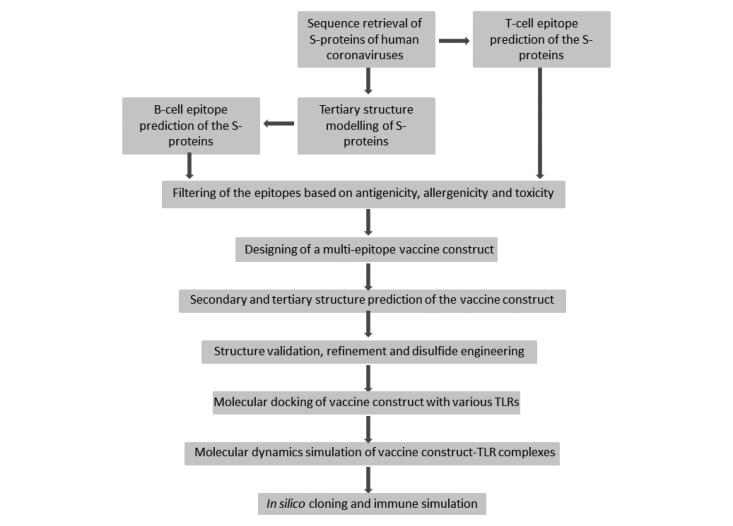
Abstract
Single stranded RNA viruses were known to cause variety of diseases since many years and are gaining much importance due to pandemic after the identification of a novel corona virus (severe acute respiratory syndrome-coronavirus (SARS-CoV-2)). Seven coronaviruses (CoVs) are known to infect humans and they are OC43 CoV, NL63 CoV, HKU1 CoV, Middle East respiratory syndrome, SARS CoV, and SARS CoV-2. Virus replication weakens the immune system of host thereby altering T-cell count and much of interferon response. Although no vaccine or therapeutic treatment has been approved till now for CoV infection, trials of vaccine against SARS CoV-2 are in progress. One of the epitopes used for vaccine production is of the spike protein on the surface of virus. The work focuses on designing of multi-epitope vaccine construct for treatment of seven human CoV infections using the epitopes present on the spike protein of human CoVs. To address this, immuno-informatics techniques have been employed to design multi-epitope vaccine construct. B- and T-cell epitopes of the spike proteins have been predicted and designed into a multi-epitope vaccine construct. The tertiary structure of the vaccine construct along with the adjuvant has been modelled and the physiochemical properties have been predicted. The multi-epitope vaccine construct has antigenic and non-allergenic property. After validation, refinement and disulphide engineering of the vaccine construct, molecular docking with toll-like receptors (TLRs) have been performed. Molecular dynamics simulation in aqueous environment predicted that the vaccine-TLRs complexes were stable. The vaccine construct is predicted to be able to trigger primary immune response in silico.
Communicated by Ramaswamy H. Sarma
Keywords: Coronavirus, spike protein, epitope, vaccine, toll-like receptor
Graphical Abstract
1. Introduction
Coronavirus (CoV) belong to large and enveloped subfamily viruses contain sense single strand RNA that are categorized into four genera namely alpha, beta, gamma, and delta, among them alpha- and beta-CoVs known to infect humans (Yi et al., 2020). Seven different types of corona virus that infect humans of alpha CoVs includes 229E and NL63; and beta-CoVs includes OC43, HKU1, MERS-CoV, and severe acute respiratory syndrome (SARS-CoV) and SARS-CoV-2 (Waleed, 2020). S, M, N, structural proteins are encoded by genome of CoV and are common for all types of virus whereas E proteins are encoded in human coronavirus 229E (HCoV-229E) and HCoV-NL63 genome, while HE protein is encoded by HCoV-OC43 and HCoV-HKU1 genome apart from structural proteins. The spike (S) protein mediates receptor-binding and fusion with the host cell membrane. The membrane (M) protein plays an important role in viral assembly. The nucleocapsid protein (N) regulates viral RNA synthesis, and may interact with M protein during virus budding. The hemagglutinin-esterase (HE) glycoprotein found only in the beta-CoVs and hemagglutinin moiety binds to neuraminic acid on the host cell surface thereby permitting initial adsorption of virus to the membrane. HCoV-229E genome consists of 27,317 bases that infects humans and bats (Lim et al., 2016). Along with HCoV-OC43, it causes common cold (Gaunt et al., 2010). HCoV-229E is associated with a range of respiratory symptoms, ranging from the common cold to pneumonia, HCoV-229E encodes four structural proteins, spike (S), envelope I, membrane (M), and nucleocapsid (N). HCoV-NL63, a member of Coronaviridae is large-envelope virus of positive-sense single-stranded RNA genome of ∼27,553 bases in length that contains a cap structure at its 5′ end and a poly (A) tail at its 3′ end. The genome consists of 5′ and 3′ noncoding regions (NCRs), and the genes encodes for four structural proteins namely spike (S), envelope I, membrane (M), and nucleocapsid (N) (Kim et al., 2018). The HCoV-OC43 genome encompasses 30,738 nucleotides of which 5′ two-thirds of the genome consists of two large replicase open reading frames (ORFs) and 3′ end includes several structural and accessory protein genes: an envelope-associated HE glycoprotein gene, a spike (S) glycoprotein gene; an envelope I protein gene; a matrix (M) glycoprotein gene; and a nucleocapsid (N) phosphoprotein gene (Vijgen et al., 2005). The genome of CoV-HKU1 is 29,926 nucleotides; polyadenylated RNA with GC content is 32%, which is lowest among all known coronaviruses. The genome organization is with the characteristic gene order 5′-replicase, spike (S), envelope I, and membrane (M), nucleocapsid (N)-3′. Both 5′ and 3′ ends contain short untranslated regions (UTRs). The 5′ end of the genome consists of a putative 5′ leader sequence (Woo et al., 2005). The first imported MERS-CoV strain was named ChinaGD01 and 30,114 nucleotide long, including the 3′ and 5′ UTRs (Lu, 2015). The genome structure is a single-stranded RNA (ssRNA) encoding replicase polyproteins (ORFs 1ab and 1a), structural proteins (E, N, and M), a surface (spike) glycoprotein (S), and sulphidral proteins (ORFs 3, 4a, 4 b, and 5) (Mackay & Arden, 2015). MERS-CoV has been assumed to be transmitted from bats and spread to humans through intermediate hosts (Coleman & Frieman, 2014). SARS genome contains 29,751 bases with 5′ cap structure and 3′ polyadenylation tract. This complex then associates with the M protein in the membranes of the ER, and virus particles form as the nucleocapsid complex buds into the lumen of the ER (Marra et al., 2003). The single-stranded RNA genome of the SARS CoV-2 was 29,891 nucleotides in size, contains two flanking UTRs and a single long ORF encoding a polyprotein, it is arranged in order of 5′-replicase (orf1/ab)-structural proteins (Spike (S)-Envelope I-Membrane (M)-Nucleocapsid (N))-3′ and lacks the HE gene (Chan et al., 2020).
No drugs or vaccines are available to treat any of the HCoV infections. Therapies to treat COVID-19 caused by SARS CoV-2 include antiviral drugs (Lu, 2020), which includes Remdesivir (Warren et al., 2016), baricitinb, interferon-α, lopinavir/ritonavir, and ribavirin (Zumla et al., 2016); immunosuppressant, steroids, and antibodies from plasma of recovered patients (Kreil & Farcet, 2018). According to WHO, hydroxychloroquine is a promising candidate to reduce the pathogenicity of SARS-CoV-2 until date.
Studies revealed that several viral structural proteins such as small envelope I, membrane (M), nucleocapsid (N), and spike (S) play important roles in SARS infection (Simmons et al., 2004). The 180-kDa spike protein plays a central role in the host cell attachment and entry processes and comprised of S1 and S2 subunits. The S1 subunit contains a signal peptide, N-terminal domain, and receptor-binding domain, while the S2 subunit contains a fusion peptide, heptad repeat (HR) 1 and 2, transmembrane domain I, and cytoplasmic domain (CP) (Chan et al., 2020). Coronavirus S proteins contain short amino-terminal hydrophobic signal sequence motifs (von Heijne, 1984). Although some coronavirus spike proteins cleaved between the S1 and S2 regions as part of activation process, unlike SARS-CoV spike not cleaved before, internalized inside the host (Xiao et al., 2003). The more variable amino-terminal region of the spike protein (S1) seems to contain the receptor-binding activity (Wong et al., 2004). The more conserved S2 region contains the transmembrane anchor, palmitic acid acylation site (Thorp et al., 2006) that is important for membrane fusion (McBride & Machamer, 2010), and the coiled-coil fusion motor domain (Bosch et al., 2003). High-resolution X-ray crystallography structures obtained for coronavirus spike protein reveals SARS-CoV in conjunction with angiotensin-I-converting enzyme 2 (ACE2) (PDB ID: 3D0G, 3D0H, 3D0I, 6ACC, 6ACD) a cellular receptor for SARS-CoV (Li et al., 2003) and HCoV-NL63 (Hofmann et al., 2005) (PDB ID: 3KBH). Image analysis of cryoelectron micrographs of SARS-CoV (Beniac et al., 2006) and other coronaviruses (Neuman et al., 2006) confirms that spikes exist as a homotrimer in the native perfusion state on virion- and protein-mediated fusion to its receptor-binding triggers conformational changes including disulphide reshuffling (Lavillette et al., 2006). Spike protein is incorporated into virion through interactions with the membrane protein M (Godeke et al., 2000). There are different kinds of receptors for different virus for effective binding to its host. 229E virus enters the host cell by binding to the aminopeptidase N receptor (Fehr & Perlman, 2015). NL63 virus enters its host cell by the ACE2 receptor (Brielle et al., 2020). OC43 virus enters its host cell by binding to the N-acetyl-9-O-acetylneuraminic acid receptor (Li, 2016). HKU1 virus enters its host cell by binding to the N-acetyl-9-O-acetylneuraminic acid receptor (Lim et al., 2016). MERS-CoV virus enters host cell by binding to the DPP4 receptor (Fehr & Perlman, 2015). SARS CoV virus infects the epithelial cells within the lungs and enters the host cell by binding to ACE2 (Ge et al., 2013). SARS CoV-2 virus enters the human cells by binding to ACE2 receptor (Letko et al., 2020).
Three HCoVs, MERS CoV, SARS CoV, and SARS CoV-2 causes severe symptoms in humans. Middle East respiratory syndrome (MERS) first reported in Saudi Arabia in September 2012 and SARS was identified in southern China in 2003. Recently, SARS CoV-2 became a pandemic spreading to 213 countries and territories. Many researchers are working towards development of vaccine for SARS CoV-2. Thus, our work focuses on designing of multi-epitope vaccine against all the HCoVs (known so far) to avoid any repeated outbreak in near future. S-protein of coronaviruses is the main agent for attachment and entry of the viruses into the human cell. The epitopes from the S-protein of seven HCoVs used to design a multi-epitope vaccine construct.
2. Materials and methods
2.1. Protein sequence retrieval
Sequences of full-length proteins of HCoV-OC43, KU1, 229E, NLC3, MERS, SARS, and novel SARS were retrieved from UniProt (http://uniprot.org) and NCBI (http://ncbi.nlm.nih.gov) databases. Sequence of OC43, KU1, 229E, NLC3, and SARS coronavirus was deposited with UniProt ID P36334, Q19U45, P15423, Q6Q1S2, and P59594, respectively. Sequence of MERS CoV and novel SARS CoV-2 was deposited in NCBI database with accession nos. AKJ80137 and QIH45053, respectively.
2.2. Multiple sequence analysis and percent identity matric analysis
Clustal Omega was used for multiple sequence analysis and phylogenetic analysis (Madeira et al., 2019). Full length sequences of HCoV-OC43, KU1, 229E, NLC3, MERS, SARS, and novel SARS were provided in FASTA format to perform the analysis. Clustal Omega uses seeded guide trees and Hidden Markov Model profile-profile technique to align two or more sequences. The phylogenetic analysis was performed with a neighbour-joining tree. Percent identity matrix was generated after phylogenetic analysis. Multiple sequence alignment was done to check the sequence similarity of the S-protein of the coronaviruses.
2.3. B-cell, CTL, and HTL epitope prediction
B-cell epitopes were predicted using the modelled tertiary structures of S-protein of HCoVs in ElliPro webserver (Ponomarenko et al., 2008). Each predicted epitope is given a score known as Protrusion Index value by the server. Maximum score is 1 and higher the score better is the predicted epitope. For each protein, a number of epitope has been predicted. For cytotoxic T-cell lymphocyte (CTL) epitope prediction NetCTL 1.2 webserver was used (Larsen et al., 2007). This server predicts the MHC-I-binding epitopes by using neural network algorithm. The server gives a score based on predicted MHC Class I-binding affinity, proteosomal C terminal cleavage and TAP transport efficiency. Higher score indicates high specificity of the epitopes towards MHC-I. The epitope with score >3 was further analysed. Helper T-cell lymphocyte (HTL) epitopes were predicted in IEDB webserver (Wang et al., 2010). The server predicted the MHC-II-binding epitopes using IEDB recommended consensus approach. Prediction of MHC-II-binding epitopes was done using HLA-DRB1 × 01:01, HLA-DPA1 × 01/DPB1 × 04:01, HLA-DQA1 × 03:01/DQB1 × 03:02 alleles. The server gives a percentile rank to the predicted epitopes. Lower rank indicates good binders. Hence, the epitopes with percentile rank of <1 were further analysed for their ability to induce IFN-γ production by using the IFNepitope server (http://crdd.osdd.net/raghava/ifnepitope/). The B-cell, CTL and IFN-γ producing HTL epitopes were further screened for antigenicity, allergenicity, and toxicity properties.
2.4. Antigenicity, allergenicity and toxicity prediction
The antigenicity of the epitopes and vaccine constructs was predicted using Vaxigen v2.0 (Doytchinova & Flower, 2007) webserver. Vaxigen v2.0 uses alignment-free approach for antigen prediction. The allergenicity of the epitopes and vaccine construct was predicted using AllerTOP v2.0 (Dimitrov et al., 2014) webserver. AllerTOP uses amino acid E-descriptors, auto- and cross-covariance transformation, and several machine learning methods for classification of allergens.
The toxicity of the epitopes was predicted using ToxinPred webserver (Gupta et al., 2013). ToxinPred can predict toxicity of peptides with <50 residues. Hence, the epitopes with more than 50 residues were chopped into fragments of 50 residues and checked for toxicity.
2.5. Construction of multi-epitope vaccine constructs
The high scoring B-cell and CTL epitopes and low scoring HTL epitopes with non-antigenic, non-allergenic and non-toxic properties were linked by appropriate linkers to design the multi-epitope construct. Also, 50S ribosomal protein L7/L12 (NCBI Accession no. P9WHE and UniProt ID P0A7K2) was used as an adjuvant and was attached to the N- terminal through EAAAK linker (Kar et al., 2020; Naz et al., 2020; Samad et al., 2020). The adjuvant is believed to improve the antigenicity of the vaccine. The B-cell and HTL epitopes were linked by GPGPG linkers and CTL epitopes were linked by AAY linkers (Kar et al., 2020; Samad et al., 2020). A 6xHis tag was incorporated at the C-terminal end to aid in protein purification and identification.
2.6. Secondary and tertiary structure prediction
Secondary structure of the vaccine construct was predicted in RaptorX webserver (Wang et al., 2011). The secondary structures are predicted in the form of helix, β-sheet and loop. Tertiary structures were predicted in I-Tasser webserver (Roy et al., 2010). I-TASSER server stands for Iterative Threading ASSEmbly Refinement server and is an integrated platform for automated protein structure and function prediction. I-TASSER first generates fragments of three-dimensional (3D) atomic models from the primary sequence by multiple threading alignments. Then the fragments are reassembled into full-length models by replica-exchange Monte Carlo simulations.
2.7. Refinement of the tertiary structure
The 3 D model obtained for the vaccine construct was refined in the 3Drefine server (http://sysbio.rnet.missouri.edu/3Drefine/). The server combines iterative optimization of hydrogen bonding network and atomic level energy minimization using composite physics and knowledge-based force fields for tertiary structure refinement (Bhattacharya et al., 2016).
2.8. Disulphide engineering
Disulphide engineering was done to rationally incorporate of disulphide bonds in the modelled structure of the vaccine construct. Disulphide engineering was done using disulphide by Design 2 v2.12 webserver (Craig & Dombkowski, 2013). The server rapidly assessed for proximity and geometry of the amino acid residues consistent with disulphide formation.
2.9. Validation of the tertiary structure
Tertiary structure model validation detects potential errors in predicted 3 D models. The structure validation was performed in SAVES v5.0 server (https://servicesn.mbi.ucla.edu/SAVES/). A Ramachandran plot was obtained in this server to visualize energetically allowed and disallowed dihedral angles psi (ψ) and phi (ϕ) of an amino acid. The server also gave ERRAT score which is used to analyse non-bonded atom-atom interactions compared to reliable high-resolution crystallography structures. Verify 3D score is also obtained from this server. This score determines the compatibility of an atomic model (3D) with its primary sequence (1D). The overall model quality was assessed in ProSA webserver (https://prosa.services.came.sbg.ac.at/prosa.php).
2.10. Physiochemical properties of the vaccine construct
ProtParam webserver (https://web.expasy.org/protparam/) was used to predict various physiochemical properties of the constructs such as amino acid composition, theoretical isoelectric point (pI), instability index, in vitro and in vivo half-life, aliphatic index, and molecular weight. Grand average of hydropathicity (GRAVY) was calculated in GRAVY Calculator webserver (http://www.gravy-calculator.de/).
2.11. Molecular docking of designed vaccine constructs with TLRs
Toll-like receptors (TLRs) are immune receptors whose activation leads to intracellular signalling pathway. TLR2 and TLR8 induce the production of NF-κB. TLR3 induces the activation of IRF3 and NF-κB. TLR4 activation leads to an intracellular signalling pathway NF-κB and inflammatory cytokine production. TLR5 induces the activation of activation leads to an intracellular signalling pathway NF-κB and IL8. All the TLRs are located at the cell membrane thus; they are the first molecules to come in contact with pathogens. Adjuvant, 50S ribosomal protein attached to the N-terminal of the vaccine construct has the ability to activate TLR4. However, other TLRs may also come in contact with the adjuvant. Thus, molecular docking of the multi-epitope vaccine construct with TLR2 (PDB ID: 6NIG), TLR3 (PDB ID: 2A0Z), TLR4 (PDB ID: 4G8A), TLR5 (PDB ID: 3J0A), and TLR8 (PDB ID: 4R0A) receptor was performed using the ClusPro 2.0 (Kozakov et al., 2017) and HDOCK (Yan et al., 2020) webservers in order to evaluate the interaction between ligand and receptor and consequently the activation of an immune response. Before docking, the X-ray crystallized structures of TLRs were prepared by removing solvent molecules and other ligands followed by addition of hydrogen atoms and energy minimization in Swiss PdbViewer Deep View software package. Binding pockets of TLRs were searched using CASTp 3.0 webserver (http://sts.bioe.uic.edu/castp/calculation.html).
2.12. Molecular dynamics simulation
The vaccine construct and vaccine construct-TLR complexes were subjected to 10 ns molecular dynamics simulation using GROMACS 4.6.5 software package (Van Der Spoel et al., 2005). For the simulation, CHARMM forcefield and SPC/E water model was used. The simulation system was neutralized by adding the suitable number of Na+/Cl– ions. The solvated system was energy minimized in 1000 steps using steepest descent method iterations. After setting the temperature at 300 K and pressure at 1 bar, production simulation was run for 30 ns to evaluate dynamics of vaccine construct and vaccine construct-TLR complexes. The RMSD, RMSF, and radius of gyration profiles of the vaccine construct and vaccine construct-TLR complexes were generated.
2.13. In silico cloning optimization of designed vaccine construct
Codon biasness occurs between prokaryotic and eukaryotic genes. Codon optimization therefore becomes necessary for protein expression in a prokaryotic host. Codon optimization of the designed multi-epitope vaccine construct was done in Java Codon Adaptation Tool (JCat) server (http://www.prodoric.de/JCat) for gene expression in the E. coli (strain K12) host. Rho-independent transcription termination, prokaryote ribosome-binding site, and restriction enzymes cleavage sites were avoided during codon optimization. Codon adaptation index (CAI) and percentage GC content was also received as output along with the optimized nucleotide sequence. The optimized nucleotide sequence of the final vaccine construct was cloned into the E. coli pET-28a (+) vector using SnapGene tool and NdeI and BglII restriction sites were introduced to the N and C-terminals of the sequence, respectively.
2.14. Immune simulation
To predict the probable immune response of the designed multi-epitope vaccine construct in human immune system, in silico immune simulations were conducted using the C-ImmSim server (http://150.146.2.1/C-IMMSIM/index.php) (Rapin et al., 2010). C-ImmSim is a novel in silico approach for the study of the mammalian immune system The tool is a combination of a mesoscopic scale simulator of the immune system with machine learning techniques for molecular-level predictions of major histocompatibility complex (MHC)–peptide-binding interactions, linear B-cell epitope discovery, and protein–protein potential estimation. All simulation parameters were kept as default and a three dose of injection (1000 antigens) at 4 weeks apart was given. The response after the injections was analysed.
3. Results
3.1. Comparison of the S-proteins of HCoVs
S-proteins of seven HCoVs causing mild and severe disorders in human have been studied. Primary structure comparison of the S-proteins revealed a sequence similarity towards the C-terminal of the proteins (Supplementary Figure 1). Based on similarity, it was found that S-protein of SARS CoV-2 and SARS CoV share the highest identity (77.46%) among all the HCoVs. S-protein of OC43 CoV and KU1 CoV share 66.21% identity and 229E CoV, and NL63 CoV share 64.07% identity. S-protein of MERS CoV shares highest identity with OC43 CoV (32.87%). The percent identity matrix of the S-proteins of HCoVs is shown in Table 1.
Table 1.
Percent identity matrix of the S-proteins of human coronaviruses.
| 229E CoV | NL63 CoV | OC43 CoV | KU1 CoV | MERS CoV | SARS CoV | SARS CoV-2 | |
|---|---|---|---|---|---|---|---|
| 229E CoV | 100 | 64.07 | 28.46 | 28.18 | 27.53 | 27.25 | 27.17 |
| NL63 CoV | 64.07 | 100 | 28.55 | 27.44 | 25.41 | 25.35 | 26.17 |
| OC43 CoV | 28.46 | 28.55 | 100 | 66.21 | 32.87 | 31.81 | 30.53 |
| KU1 CoV | 28.18 | 27.44 | 66.21 | 100 | 32.50 | 31.05 | 30.15 |
| MERS CoV | 27.53 | 25.41 | 32.87 | 32.50 | 100 | 32.27 | 31.93 |
| SARS CoV | 27.25 | 25.35 | 31.81 | 31.05 | 32.27 | 100 | 77.46 |
| SARS CoV-2 | 27.17 | 26.17 | 30.53 | 30.15 | 31.93 | 77.46 | 100 |
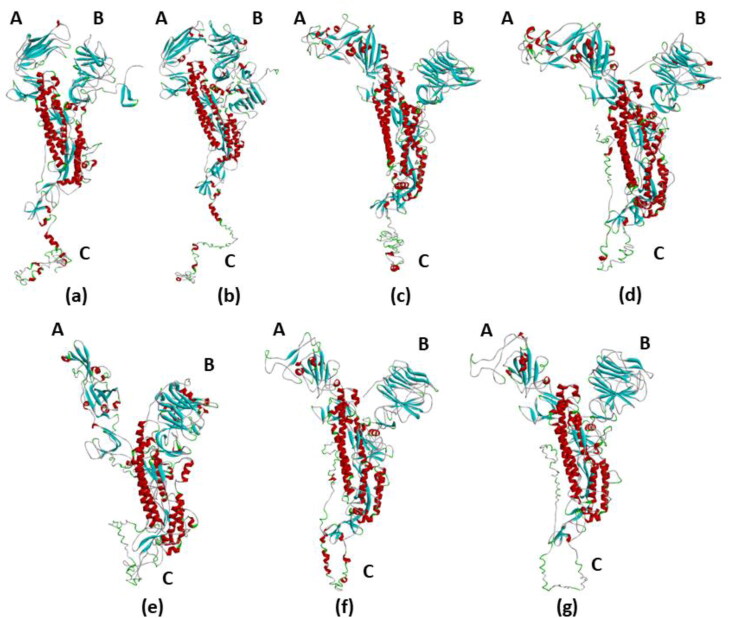
Tertiary structure of full-length S-proteins of HCoVs shows a similar Y-shaped structure. Arm A is the receptor-binding domain. Arm C contains the transmembrane domain and the cytoplasmic domain. From Figure 1, it is clearly seen that the arms A and B of S-protein of 229E CoV and NL63 CoV is in closed conformation. The arms A and B of S-proteins of all other HCoVs are in open conformation. Another difference in the tertiary structure is seen in the C arm, which is the C-terminal tail of S-proteins. The C arm of S-protein of 229E CoV and NL63 CoV is in an elongated form, whereas the C arm is in a loop like structure in the other HCoVs.
Figure 1.
Tertiary structures of the S-protein of human coronaviruses. The S-proteins have a similar Y-shaped tertiary structure. The three arms of the S-protein is represented by A, B, and C. Arm A is the receptor-binding domain and arm C is the C-terminal transmembrane and cytoplasmic domain. Red colour represents α-helix, blue colour represents β-sheets, green colour represents turns, and white colour represents coils and loops. (a) 229E CoV, (b) NL63 CoV, (c) OC43 CoV, (d) KU1 CoV, (e) MERS CoV, (f) SARS CoV, and (g) SARS CoV-2.
3.2. B-cell CTL and HTL epitope prediction
B-cell epitopes of different lengths were predicted for each S-protein of HCoVs (Supplementary Tables 1–7). CTL epitopes of 9 residues along with their MHC-binding affinity, C-terminal cleavage affinity and transport affinity were predicted (Supplementary Table 8). HTL epitopes of 15 residue length interacting with HLA-DRB1 × 01:01 and HLA-DQA1 × 01:01/DQB1 × 06:01 alleles were predicted (Supplementary Tables 9–13). The epitopes were subjected to allergenicity, antigenicity and toxicity prediction. The epitopes with best scores and non-allergenic, antigenic, and non-toxic properties were selected for the vaccine construct.
3.3. Designing of multi-epitope vaccine construct
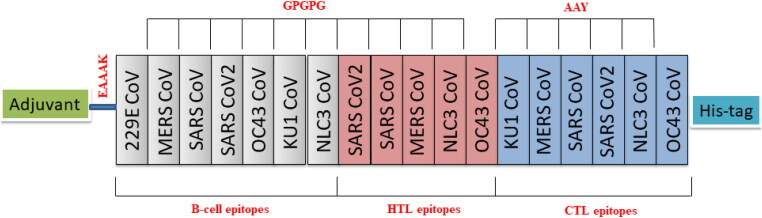
Eighteen epitopes from the S-protein of HCoVs were linked by linkers to form the final vaccine construct (Table 2). The B-cell and HTL epitopes were linked by GPGPG linkers whereas the CTL epitopes were linked by AAY linkers. The TLR4 agonist, 50S ribosomal L7/L12 protein was added to the amino terminus of the vaccine construct using an EAAAK linker and 6xHis tag was added at the C-terminal to help in protein purification and identification. The final vaccine construct designed consisted of 1189 amino acid residues (Figure 2).
Table 2.
Epitopes of the S-protein of human coronaviruses selected for final multi-epitope vaccine construct.
| Epitope type | Coronavirus S-protein | Start residue | End residue | Epitope sequence |
|---|---|---|---|---|
| B-cell epitope | 229E | 1 | 42 | MFVLLVAYALLHIAGCQTTNGLNTSYSVCNGCVGYSENVFAV |
| OC43 | 437 | 590 | LPAANVSVSRFNPSTWNKRFGFIEDSVFKPRPAGVLTNHDVVYAQHCFKAPKNFCPCKLNGSCVGSGPGKNNGIGTCPAGTNY LTCDNLCTPDPITFTGTYKCPQTKSLVGIGEHCSGLAVKSDYCGGNSCTCRPQAFLGWSADSCLQGDKCNI | |
| KU1 | 1151 | 1293 | TSFKTVLVSPGLCLSGDRGIAPKQGYFIKQNDSWMFTGSSYYYPEPISDKNVVFMNSCSVNFTKAPFIYLNNSIPNLSDFEAEF SLWFKNHTSIAPNLTFNSHINATFLDLYYEMNVIQESIKSLNSSFINLKEIGTYEMYVK | |
| NLC3 | 1212 | 1356 | PDYVDVNKTLQEFAQNLPKYVKPNFDLTPFNLTYLNLSSELKQLEAKTASLFQTTVELQGLIDQINSTYVDLKLLNRFENYIK WPWWVWLIISVVFVVLLSLLVFCCLSTGCCGCCNCLTSSMRGCCDCGSTKLPYYEFEKVHVQ | |
| MERS | 172 | 251 | LPDGCGTLLRAFYCILEPRSGNHCPAGNSYTSFATYHTPATDCSDGNYNRNASLNSFKEYFNLRNCTFMYTYNITEDEIL | |
| SARS CoV | 377 | 522 | LCFSNVYADSFVVKGDDVRQIAPGQTGVIADYNYKLPDDFMGCVLAWNTRNIDATSTGNYNYKYRYLRHGKLRPFERDIS NVPFSPDGKPCTPPALNCYWPLNDYGFYTTTGIGYQPYRVVVLSFELLNAPATVCGPKLSTDLIKN | |
| SARS CoV-2 | 390 | 536 | LCFTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQ AGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCGPKKSTNLVKN | |
| CTL epitopes | OC43 | 953 | 961 | LSENQISGY |
| KU1 | 248 | 256 | NTDNETLEY | |
| NLC3 | 1272 | 1278 | LQINSTY | |
| MERS | 769 | 777 | QVDQLNSSY | |
| SARS CoV | 847 | 855 | LTDDMIAAY | |
| SARS CoV-2 | 865 | 873 | LTDEMIAQY | |
| HTL epitopes | OC43 | 972 | 986 | PPWTAAAGVPFYLNV |
| NLC3 | 940 | 954 | GGLTSAAAIPFSLAL | |
| MERS | 959 | 973 | GWTAGLSSFAAIPFA | |
| SARS CoV | 852 | 866 | IAAYTAALVSGTATA | |
| SARS CoV-2 | 512 | 526 | VLSFELLHAPATVCG |
Figure 2.
Schematic diagram of final multi-epitope vaccine construct. The multi-epitope construct sequence consisted of adjuvant followed by epitopes and His-tag. Adjuvant is linked to the epitope by EAAAK linker. B-cell epitope and HTL epitopes are inter-linked by GPGPG linker and CTL epitopes are linked by AAY linkers.
3.4. Antigenicity and allergenicity prediction of the vaccine construct
The antigenicity of the vaccine construct including the adjuvant sequence and His-tag was predicted by the VaxiJen 2.0 server to be 0.6452 with a bacteria model at a threshold of 0.4. The main vaccine sequence without adjuvant and His-tag gave scores of 0.6635 with bacteria model at a threshold of 0.4 on VaxiJen 2.0 server. Thus, the results indicate that the designed vaccine is antigenic in nature. The presence of adjuvant and His-tag doesn’t change the antigenic property of the construct. The vaccine construct with and without the adjuvant and His-tag is predicted to be non-allergenic by AllerTOP v.2 and AllergenFP servers.
3.5. Physiochemical properties of the vaccine construct
The calculated molecular weight of the vaccine construct is 128.381 kDa. Theoretical pI value of the construct is 5.57, indicating the protein is slightly acidic in nature. The estimated half-life of the vaccine construct is 30 h mammalian reticulocytes in vitro, and >20 h in yeast in vivo and >10 h in E. coli in vivo. The instability index (II) is computed to be 29.71, which classify the protein as stable. The aliphatic index is calculated to be 29.71 and grand average of hydropathicity (GRAVY) score is predicted to be −0.072, indicating that protein is hydrophilic in nature.
3.6. Secondary and tertiary structure prediction of the vaccine constructs
The secondary structure of the multi-epitope vaccine construct with adjuvant and His-tag is predicted to be composed of 17% α-helix, 17% β-sheet, and 64% coils. Of the total residues 42% are predicted to be exposed and 29% residues are predicted to be buried. Tertiary structure prediction by I-TASSER server predicted five tertiary structure models of the peptide aptamer. Ten threading templates were used for the prediction, of which 6jx7A, 1rquA, 6nzkA, and 5ijoJ were the best templates. All the templates showed good alignment as per their Z-score values, ranging from 1.40 to 18.24. The calculated C-score of the five predicted models ranges from −1.49 to −2.69. The C-score range is generally between −5 and 2 and higher value indicates higher confidence (Lee et al., 2010). Thus, the model with the highest C-score was selected for further studies. This model had a TM-score of 0.51 ± 0.15 with an estimated RMSD of 13.6 ± 4.0 Å. The TM-score is a measure of structural similarity between two structures (Shaik, 2019). A TM-score of >0.5 indicates a model of correct topology and a TM score <0.17 means random similarity (Shaik, 2019). These cut-off values are independent of the length of protein.
3.7. Validation, refinement, and disulphide engineering of the vaccine construct
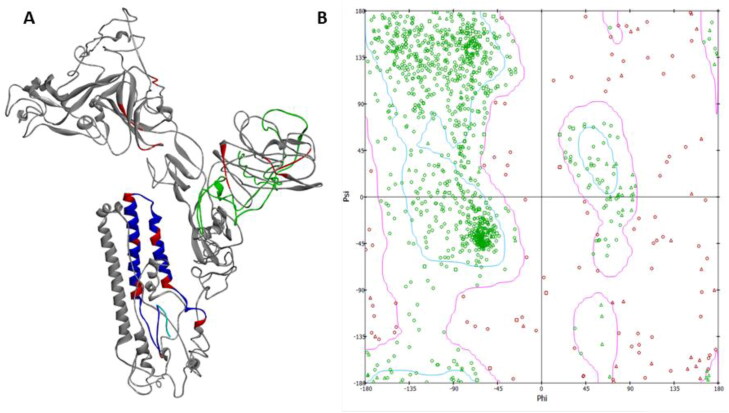
The tertiary structure of the vaccine construct was refined on 3Drefine server and it yielded five models. Model 1 was found to be the best model based on model quality scores for all refined models (Figure 3(A)). The model quality scores included high accuracy global distance test (GDT-HA) score of 0.9899, global distance test total score of 0.9899, RMSD score of 0.263 Å, MolProbity score of 3.835, RWplus score of −201,345.267, and 3Drefine score of 11,651.6. This model was chosen as the final vaccine construct model for further analysis.
Figure 3.
(A) Tertiary structure of the multi-epitope vaccine construct. Green colour depicts the N- terminal adjuvant. Grey colour refers to B-cell and HTL epitopes. Blue colour refers to CTL epitopes. Linkers are represented by red colour. (B) Ramachandran plot of the modelled structure generated in Biovia Discovery studio.
Disulphide engineering was performed with 10 selected pair of residues (Pro563-Asn566, Gly44-Ala135, Gly75-Gly80, Pro786-Leu829, Glu29-Phe31, Glu113-Ala116, Gln143-Cys 239, Phe1109-Try1110, Leu553-Asn556, and Ala1053-Phe1072). The selection was done on the basis of energy, χ3 value, and B-factor. The residue pairs with energy of 1 kcal/mol to 2.2 kcal/mol (mean value is 1.0 kcal/mol, and the 90th percentile is 2.2 kcal/mol), χ3 value of −85° to −90° and +95° to +100° (mean value is −87° and +97°) and B-factor of 29–33 (mean B-factor for residues involved in stabilizing disulphide bonds is 31.6) (Craig & Dombkowski, 2013).
After refinement and disulphide engineering of the modelled structure, validation was done. The overall model quality was −2 as calculated by ProSA. The Ramachandran plot generated showed that 74.2% of the residues are in favourable region, 15.2% residues are in the allowed region, and 10.6% residues are in outline region. The quality and potential errors in the tertiary structure were verified by ERRAT. The chosen model after refinement had an overall quality factor of 64.91% with ERRAT. Verify3D also passed the modelled structure as >80% of the residues have averaged 3 D-1D score ≥0.2. Verify3D determines the compatibility of an atomic model (3D) with its own amino acid sequence (1D).
3.8. Molecular docking of vaccine construct and TLRs
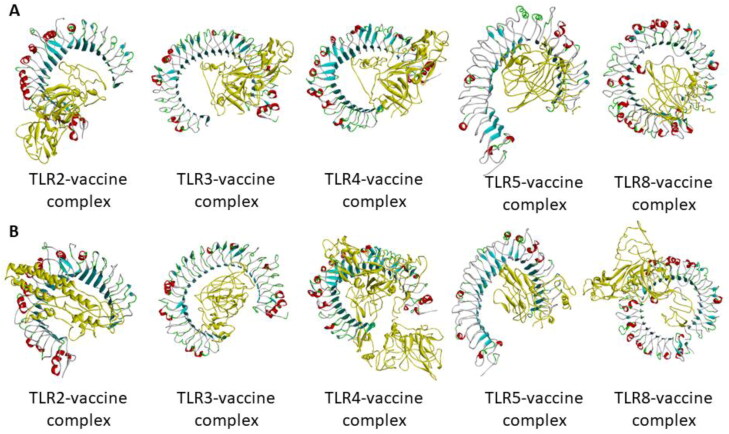
Binding pocket prediction of the studied TLRs revealed one pocket. However, the solvent accessible surface area and volume of the binding pocket of all the TLRs are not similar (Table 3). Molecular docking was performed in HDOCK server and ClusPro 2.0 server. HDOCK server predicted 100 complexes in each case from which the lowest scoring complex was studied. ClusPro 2.0 predicted 10 complexes from which the lowest energy complex was studied. From Figure 4, it can be seen that the vaccine construct has almost similar binding pattern to TLR3, TLR4, and TLR5. The adjuvant containing arm of the vaccine could interact with the predicted binding pocket of the TLRs. Based on binding energy, TLR5-vaccine construct complex is predicted to be the best complex in both the servers (Table 3). Thus, although the adjuvant used is a known TLR4 agonist, the vaccine construct may have affinity towards TLR3 and TLR5 as well.
Table 3.
Predicted solvent accessible surface area and volume of the binding pocket of TLRs and binding energies upon interacting with the vaccine construct.
| TLR | No. of binding pockets | Solvent accessible surface area | Solvent accessible volume | Binding energy upon complex formation with TLRs (kcal/mol) |
|
|---|---|---|---|---|---|
| HDOCK server | ClusPro 2.0 server | ||||
| TLR2 | 1 | 2973.383 | 20,582.911 | –299 | –1386.6 |
| TLR3 | 1 | 2973.383 | 20,582.911 | –299.5 | –1538.4 |
| TLR4 | 1 | 1905.100 | 11,916.551 | –310.49 | –1341.5 |
| TLR5 | 1 | 2925.540 | 7490.737 | –371.20 | –1646.5 |
| TLR8 | 1 | 2925.540 | 7490.737 | –304.11 | –1260.8 |
Figure 4.
Molecular docking of vaccine construct with TLR2, TLR3, TLR4, TLR5, and TLR8 by two servers: (A) HDOCK server and (B) ClusPro 2.0 server. The vaccine construct is represented in yellow colour.
3.9. Molecular dynamics simulation
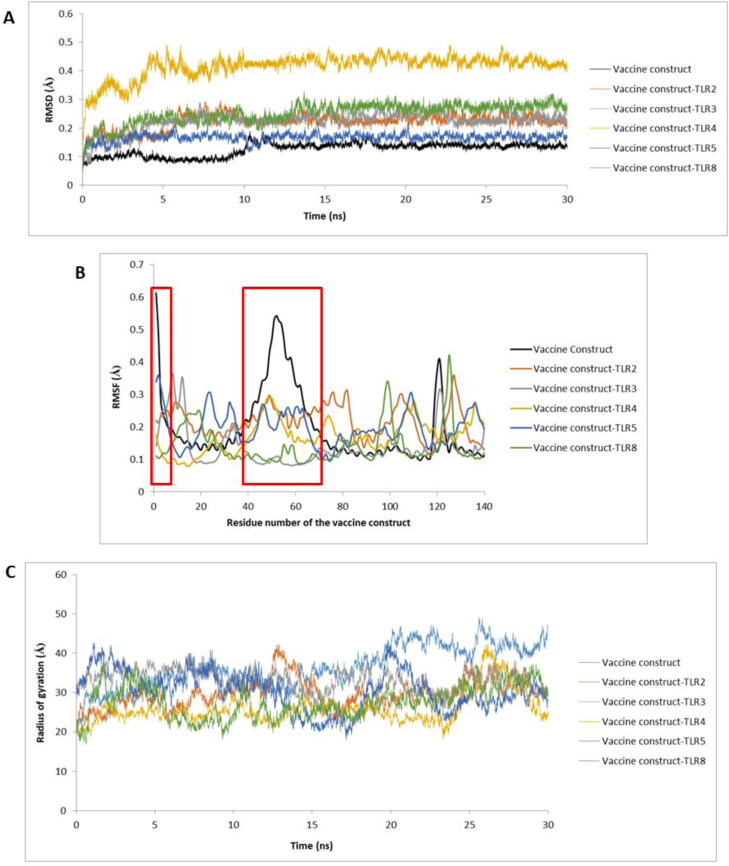
Molecular dynamics simulation of the vaccine construct revealed its stability throughout the simulation. The average RMSD of the vaccine construct alone was 0.08 Å. Upon binding to TLRs, the average RMSD of the vaccine construct increased. The vaccine construct-TLR complexes were found to be stable with no major fluctuation (Figure 5(A)). The vaccine construct has shown a fluctuation at 12 ns but was stable after that. One major fluctuation in the vaccine construct-TLR4 complex was seen at 3 ns. The complex was found to be stable after that. The average RMSD of this complex was 0.5 Å which is higher than all other complexes. The RMSF of vaccine construct showed that the N-terminal residues and the residues between 40 and 65 positions have highest movement (Figure 5(B)). The N-terminal of the vaccine construct (residue 1–121) consists of the adjuvant and is involved in interaction with TLRs. However, when the vaccine construct is bound to the TLRs, the RMSF of the highest fluctuating residues of the construct has decreased. This can be due to interaction with TLRs. Radius of gyration of vaccine construct after 30 ns simulation was calculated to be 45.74 nm. However, the radius of gyration of vaccine-construct-TLR complexes ranged between 29.47 and 35.67 nm. The radius of gyration profile against time (in ns) reveals that the vaccine construct alone and vaccine construct-TLR complexes were in a compact state till the end of molecular dynamics run (Figure 5(C)).
Figure 5.
(A) RMSD profile, (B) RMSF profile, and (C) Radius of gyration profile of vaccine construct and vaccine construct-TLR complexes. The RMSF profile of first 140 residues of the vaccine construct is shown here as this region consists of the adjuvant which is involved in interacting with TLRs.
3.10. Codon optimization of the final vaccine construct
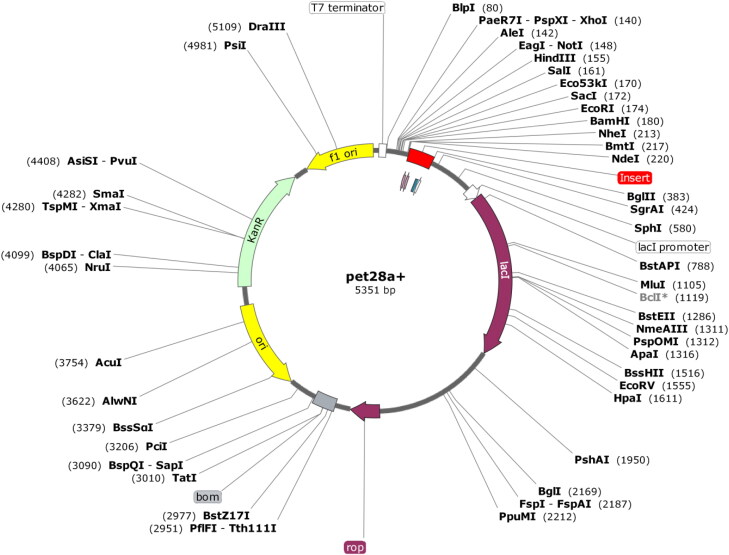
Codon optimization of the vaccine construct in E. coli (strain K12) was done for expression of protein. The optimized codon sequence was 3266 nucleotides long. The CAI of the optimized nucleotide sequence was 1.0 which means that the nucleotide sequence contains the most frequently occurring codons (Rapin et al., 2010). The average GC content of the optimized sequence is 52.20%. Higher the GC content, better is the expression of a protein in prokaryotes (Zhou et al., 2014; Du, 2018). The optimized nucleotide sequence was inserted into the pET28a (+) vector using SnapGene software (Figure 6).
Figure 6.
In silico restriction cloning of final multi-epitope vaccine construct into pET28a(+) expression vector. Red part represents the vaccine insert.
3.11. Immune simulation
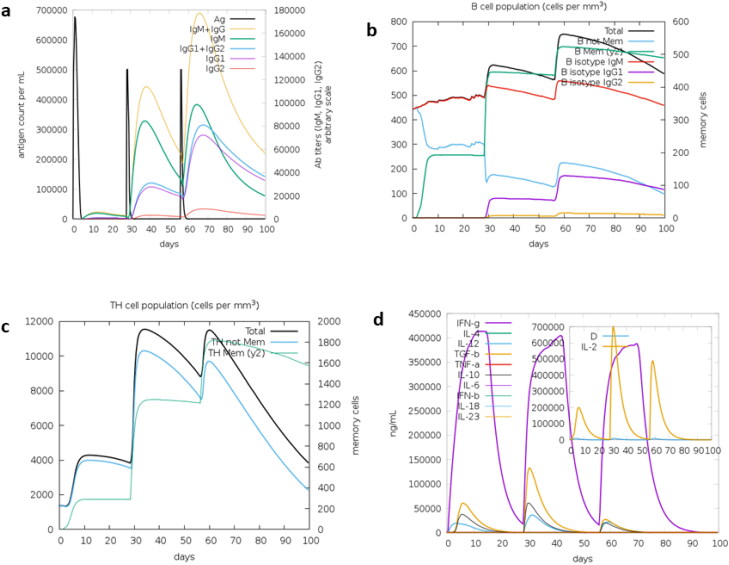
Immune simulation by C-ImmSim server showed the immune responses of three injections. The antigen is calculated to be present for about 5 days after each dose. After first dose, very low level of IgM is seen to be present. But after second and third dose, the level of IgM and IgG subsequently increased. After third dose, the total level of IgM and IgG antibodies is found to be higher than the level of antigen. Similarly, level of memory B-cell is seen to be increasing after each dose. The level of memory T-cells also increased after second dose. However, after third dose, there was no further increase in the level of memory T-cells. The level of IFN-γ was found to be increasing at the same level after each dose. The level of IFN-γ dropped to basal level at the end of fourth week and subsequent injection of the antigen again triggered the expression of IFN-γ. Another cytokine IL-2 was found to reach its maximum expression after the second dose. The level of IgG, IgM, and cytokines dropped gradually after 4 weeks but the level of memory B-cells and memory T-cells were found to be constant. Thus, the vaccine construct is predicted to trigger mostly IgG, IgM, B-cell, T-cell, and cytokines (IFN-γ and IL2) (Figure 7).
Figure 7.
C-ImmSim presentation of an in silico immune simulation with the multi-epitope vaccine construct. (a) Immunoglobulin production in response to antigen injection, (b) B-cell populations after antigen injection, (c) production of T-helper cells, and (d) cytokine production after the injection.
4. Discussion
Three HCoV s are known to cause severe respiratory symptoms. MERS CoV outbreak was seen in 2003, SARS CoV became epidemic after affecting 26 countries in 2012 and SARS CoV-2 pandemic started towards the end of 2019. Other HCoVs, 229E, NLC3, OC43, and KU1 causes mild respiratory symptoms but 229E and KU1 is also known to cause pneumonia. Until the outbreak of SARS CoV-2, there is no approved treatment for any of the HCoV infection. After the outbreak of SARS CoV-2, researchers are trying to develop vaccine against SARS CoV-2. B-cell, CTL, and HTL epitopes are used as antigenic determinants for efficient vaccine production had much focused on spike (S) protein, which is gaining much attention in research. Here in this paper we had focused on antigenic determinants on the S-protein of HCoVs and their use as effective vaccine candidates through in silico approach. Sequence comparison of S-proteins of human corona viruses shows similarity towards the C-terminal end. However, tertiary structure shows a similar Y-shaped conformation of the S-proteins. To get insights and details about various antigenic determinants of S-proteins of virus different lengths of B-cell epitopes were predicted using ElliPro server. CTL epitopes were predicted using NetCTL 1.2 server and HTL epitopes were predicted using IEDB server. The HTL epitopes were checked for their ability to produce IFN-γ using IFN epitope server. HTL epitopes predicted for HCoVs 229E and KU1 didn’t have the ability to produce IFN-γ. All the predicted epitopes were further checked for allergenic, immunogenic, and toxic properties. The high-scoring epitopes (as per the servers) with antigenic, non-allergenic, and non-toxic properties were selected for vaccine construct designing. Multi-epitope vaccine construct was designed by linking the selected B-cell and HTL epitopes with GPGPG linkage. The CTL epitopes were linked by AAY linkers. 50S ribosomal protein, which is a TLR4 agonist is used as an adjuvant and linked to the N-terminus with EAAAK linkage .Also, for ease of protein purification, 6× His tag is added at C-terminus, thus yielding a final construct of 1189 amino acids. Immunogenic properties such as antigenic and allergic predictions performed using VaxiJen v2.0 and AllerTOP v.2 server indicates designed vaccine construct is antigenic and non-allergic in nature.
The molecular weight of the designed multi-epitope vaccine construct is found to be 128.381 kDa with pI of 5.57 and a half-life of 30 h in mammalian reticulocytes. The vaccine construct is also predicted to be stable and hydrophilic in nature. Secondary structure prediction of multi-epitope vaccine construct as found to have 17% α-helix, 17% β-sheet, and 64% coils. Of the total residues, 42% are predicted to be exposed and 29% residues are predicted to be buried. The tertiary structure of the vaccine construct was refined on 3Drefine server. The refined model GDT-HA score of 0.9899, global distance test-total score of 0.9899, RMSD score of 0.263 Å, MolProbity score of 3.835, RWplus score of −201,345.267, and 3Drefine score of 11,651.6. Disulphide engineering was performed with ten selected pair of residues (Pro563-Asn566, Gly44-Ala135, Gly75-Gly80, Pro786-Leu829, Glu29-Phe31, Glu113-Ala116, Gln143-Cys 239, Phe1109-Try1110, Leu553-Asn556, and Ala1053-Phe1072). The selection was done on the basis of energy, ꭓ3 value, and B-factor. After refinement and disulphide engineering of the modelled structure, validation was done. The overall model quality was −2 as calculated by ProSA. The Ramachandran plot generated showed that 74.2% of the residues are in favourable region, 15.2% residues are in the allowed region and 10.6% residues are in outline region. The model had an overall quality factor of 64.91% with ERRAT. Verify3D also passed the modelled structure as more than 80% of the residues have averaged 3D-1D score ≥0.2.
Molecular docking was performed in HDOCK server and ClusPro 2.0 server. The vaccine construct has almost similar binding pattern to TLR3, TLR4, and TLR5. The adjuvant containing arm of the vaccine could interact with the predicted binding pocket of the TLRs. Based on binding energy, TLR5-vaccine construct complex is predicted to be the best complex in both the servers. The stability of the TLR-vaccine construct complexes were checked by performing a 30 ns molecular dynamics simulation in GROMACS 4.6.5 software package. CHARMM force field and SPC/E water model was chosen for molecular dynamics simulation. The average RMSD of the vaccine construct alone was 0.08 Å. Upon binding to TLRs, the average RMSD of the vaccine construct increased. The vaccine construct-TLR complexes were found to be stable. TLR4-vaccine construct complex showed fluctuation at 3 ns. After 3 ns, there was no major fluctuation in the RMSD profile of TLR4-vaccine construct complex. RMSF profile of the vaccine construct showed the presence of two highly fluctuating regions. The regions are the N-terminal region and the region between 40 and 65 positions. The N-terminal of the vaccine construct (residues 1–121) consists of the adjuvant and is involved in interaction with TLRs. Interaction of the vaccine construct with TLRs have decreased the fluctuations of the two regions. Radius of gyration of vaccine construct after 30 ns simulation was calculated to be 45.74 nm. However, the radius of gyration of vaccine-construct-TLR complexes ranged between 29.47 and 35.67 nm. This change in radius of gyration reveals the compactness of the vaccine construct-TLR complexes.
The vaccine construct must be cloned into a suitable vector for expression and purification. pET28a(+) is a suitable vector for protein expression. Prokaryotic vectors often have codon biasness while expressing eukaryotic proteins. Thus, codon optimization was done to avoid codon biasness. The CAI of the optimized nucleotide sequence was 1.0 and average GC content of the optimized sequence is 52.20%. These two parameters show that the vaccine construct will be highly expressed in the vector. In silico prediction of primary immune response after vaccine injection was also done in C-ImmSim server. Three injections of 1000 antigen molecules without LPS were given 4 weeks apart and immune response was recorded for 100 days. The antigen was seen to be present for about 5 days after each dose. After first dose, very low level of IgM was generated. But after second and third dose, the level of IgM and IgG subsequently increased. After third dose, the total level of IgM and IgG antibodies is found to be higher than the level of antigen. Similarly, level of memory B-cell increased after each dose. The level of memory T-cells also increased after second dose. However, after third dose, there was no further increase in the level of memory T-cells. The level of IFN-γ was found to be increasing at the same level after each dose. The level of IFN-γ dropped to basal level at the end of fourth week and subsequent injection of the antigen again triggered the expression of IFN-γ. Another cytokine IL-2 was found to reach its maximum expression after the second dose. The level of their cytokines such as IL4, IL12, IL10, IL6, IL18, IFN-α, TGF-β, and IFN-β did not undergo much change even after three doses of antigen injection. The level of IgG, IgM, and cytokines dropped gradually after 4 weeks but the level of memory B-cells and memory T-cells were found to be constant. Thus, the vaccine construct is predicted to trigger mostly IgG, IgM, B-cell, T-cell, and cytokines (IFN-γ and IL2).
5. Conclusion
Complete eradication of a disease or infection is not possible without a specific treatment. There is always a possibility that previous epidemics can re-emerge and cause severe damage to coming generations. The outbreak of MERS and SARS CoV caused severe damage to some countries but till date no vaccine or treatment has been approved. The present outbreak of SARS CoV-2 has triggered the importance of vaccine. Thus, in this work, we have employed immune-informatics tools to design multi-epitope vaccine construct against the S-protein of seven HCoVs. The S-protein is the medium by which the viral genome transfers to human cells. Thus, the vaccine construct designed is expected to trigger host immune system to generate antibodies against the S-protein epitope and restrict the entry of viral genome to host cells. This multi-epitope vaccine may help in boosting the host immune system against seven HCoVs.
Supplementary Material
Acknowledgements
A.D. acknowledges CSIR-SRF for providing financial assistance. N.S.N.C. acknowledges ICMR-SRF for proving financial assistance. The authors acknowledge the anonymous reviewers for helping to improve the manuscript by providing critical suggestions.
Glossary
Abbreviations
- MERS
Middle East respiratory syndrome
- SARS
Severe acute respiratory syndrome
- CoV
Coronavirus
- DPP4
Dipeptidyl-peptidase 4
- ACE 2
Angiotensin-converting enzyme 2
- CTL
Cytotoxic T-cell lymphocyte
- HTL
Helper T-cell lymphocyte
- IL
Interleukin
- TNF
Tumour necrosis factor
- B-cell
Bone marrow cell
- T-cell
Thymus cell
- HLA
Human leukocyte antigen
- PDB
Protein data bank
- I-TASSER
Iterative Threading ASSEmbly Refinement
- TLR
Toll-like receptor
- GROMACS
GROningen MAchine for Chemical Simulations
- CHARMM,
Chemistry at Harvard macromolecular mechanics
- SPC/E
Extended simple point charge
- TM-score
Template modelling score
- RMSD
Root mean square deviation
- RMSF
Root mean square fluctuation
- Ig
Immunoglobulin
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Beniac, D. R., Andonov, A., Grudeski, E., & Booth, T. F. (2006). Architecture of the SARS coronavirus prefusion spike. Nature Structural and Molecular Biology, 13(8), 751–752. 10.1038/nsmb1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya, D., Nowotny, J., Cao, R., & Cheng, J. (2016). 3Drefine: An interactive web server for efficient protein structure refinement. Nucleic Acids Research, 44(W1), W406–409. 10.1093/nar/gkw336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch, B. J., van der Zee, R., de Haan, C. A. M., & Rottier, P. J. M. (2003). The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. Journal of Virology, 77(16), 8801–8811. 10.1128/jvi.77.16.8801-8811.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brielle, E. S., Schneidman-Duhovny, D., & Linial, M. (2020). The SARS-CoV-2 exerts a distinctive strategy for interacting with the ACE2 human receptor. Viruses, 12(5), 497. 10.3390/v12050497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J. F.-W., Kok, K.-H., Zhu, Z., Chu, H., To, K. K.-W., Yuan, S., & Yuen, K.-Y. (2020). Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerging Microbes and Infections, 9(1), 221–236. 10.1080/22221751.2020.1719902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, C. M., & Frieman, M. B. (2014). Coronaviruses: Important emerging human pathogens. Journal of Virology, 88(10), 5209–5212. 10.1128/JVI.03488-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, D. B., & Dombkowski, A. A. (2013). Disulfide by Design 2.0: A web-based tool for disulfide engineering in proteins. BMC Bioinformatics, 14, 346. 10.1186/1471-2105-14-346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov, I., Bangov, I., Flower, D. R., & Doytchinova, I. (2014). AllerTOP v.2-a server for in silico prediction of allergens. Journal of Molecular Modeling, 20(6), 2278. 10.1007/s00894-014-2278-5 [DOI] [PubMed] [Google Scholar]
- Doytchinova, I. A., & Flower, D. R. (2007). VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics, 8(1), 4. 10.1186/1471-2105-8-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, M. Z. (2018). The GC content as a main factor shaping the amino acid usage during bacterial evolution process. Frontiers in Microbiology, 9, 2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr, A. R., & Perlman, S. (2015). Coronaviruses: An overview of their replication and pathogenesis. Methods in Molecular Biology, 1282, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt, E. R., Hardie, A., Claas, E. C. J., Simmonds, P., & Templeton, K. E. (2010). Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. Journal of Clinical Microbiology, 48(8), 2940–2947. 10.1128/JCM.00636-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, X.-Y., Li, J.-L., Yang, X.-L., Chmura, A. A., Zhu, G., Epstein, J. H., Mazet, J. K., Hu, B., Zhang, W., Peng, C., Zhang, Y.-J., Luo, C.-M., Tan, B., Wang, N., Zhu, Y., Crameri, G., Zhang, S.-Y., Wang, L.-F., Daszak, P., & Shi, Z.-L. (2013). Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature, 503(7477), 535–538. 10.1038/nature12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godeke, G. J., de Haan, C. A., Rossen, J. W., Vennema, H., & Rottier, P. J. (2000). Assembly of spikes into coronavirus particles is mediated by the carboxy-terminal domain of the spike protein. Journal of Virology, 74(3), 1566–1571. 10.1128/jvi.74.3.1566-1571.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S., Kapoor, P., Chaudhary, K., Gautam, A., Kumar, R., & Raghava, G. P. S, Open Source Drug Discovery Consortium (2013). In silico approach for predicting toxicity of peptides and proteins. PLoS One, 8(9), e73957. 10.1371/journal.pone.0073957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, H., Pyrc, K., van der Hoek, L., Geier, M., Berkhout, B., & Pöhlmann, S. (2005). Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proceedings of the National Academy of Sciences of the United States of America, 102(22), 7988–7993. 10.1073/pnas.0409465102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar, T., Narsaria, U., Basak, S., Deb, D., Castiglione, F., Mueller, D. M., & Srivastava, A. P. (2020). A candidate multi-epitope vaccine against SARS-CoV-2. Scientific Reports, 10(1), 10895. 10.1038/s41598-020-67749-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.-M., Kim, S.-T., Yang, J.-S., Kim, S. S., & Cheong, H.-M. (2018). Complete genome sequence of human coronavirus NL63 CN0601/14, first isolated in South Korea. Genome Announcements, 6(12), e00152–18. 10.1128/genomeA.00152-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov, D., Hall, D. R., Xia, B., Porter, K. A., Padhorny, D., Yueh, C., Beglov, D., & Vajda, S. (2017). The ClusPro web server for protein-protein docking. Nature Protocols, 12(2), 255–278. 10.1038/nprot.2016.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreil, T. R., & Farcet, M. R. (2018). Immunoglobulins and virus antibody titers: Of past needs, current requirements, and future options. Transfusion, 58(Suppl 3), 3090–3095. 10.1111/trf.15016 [DOI] [PubMed] [Google Scholar]
- Larsen, M. V., Lundegaard, C., Lamberth, K., Buus, S., Lund, O., & Nielsen, M. (2007). Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics, 8, 424. 10.1186/1471-2105-8-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavillette, D., Barbouche, R., Yao, Y., Boson, B., Cosset, F.-L., Jones, I. M., & Fenouillet, E. (2006). Significant redox insensitivity of the functions of the SARS-CoV spike glycoprotein: Comparison with HIV envelope . Journal of Biological Chemistry, 281(14), 9200–9204. 10.1074/jbc.M512529200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Weon, S., Lee, S., & Kang, C. (2010). Relative codon adaptation index, a sensitive measure of codon usage bias. Evolutionary Bioinformatics, 6, 47–55. 10.4137/EBO.S4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko, M., Marzi, A., & Munster, V. (2020). Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature Microbiology, 5(4), 562–569. 10.1038/s41564-020-0688-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. (2016). Structure, function, and evolution of coronavirus spike proteins. Annual Review of Virology, 3(1), 237–261. 10.1146/annurev-virology-110615-042301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., Somasundaran, M., Sullivan, J. L., Luzuriaga, K., Greenough, T. C., Choe, H., & Farzan, M. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature, 426(6965), 450–454. 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, Y., Ng, Y., Tam, J., & Liu, D. (2016). Human coronaviruses: A review of virus-host interactions. Diseases, 4(4), 26. 10.3390/diseases4030026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H. (2020). Drug treatment options for the 2019-new coronavirus (2019-nCoV). Bioscience Trends, 14(1), 69–71. 10.5582/bst.2020.01020 [DOI] [PubMed] [Google Scholar]
- Lu, R. (2015). Complete genome sequence of Middle East respiratory syndrome coronavirus (MERS-CoV) from the first imported MERS-CoV case in China. Genome Announcements, 3(4):e00818–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, I. M., & Arden, K. E. (2015). Middle East respiratory syndrome: An emerging coronavirus infection tracked by the crowd. Virus Research, 202, 60–88. 10.1016/j.virusres.2015.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., Basutkar, P., Tivey, A. R. N., Potter, S. C., Finn, R. D., & Lopez, R. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Research, 47(W1), W636–w641. 10.1093/nar/gkz268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, M. A., Jones, S. J. M., Astell, C. R., Holt, R. A., Brooks-Wilson, A., Butterfield, Y. S. N., Khattra, J., Asano, J. K., Barber, S. A., Chan, S. Y., Cloutier, A., Coughlin, S. M., Freeman, D., Girn, N., Griffith, O. L., Leach, S. R., Mayo, M., McDonald, H., Montgomery, S. B., … Roper, R. L. (2003). The Genome sequence of the SARS-associated coronavirus. Science, 300(5624), 1399–1404. 10.1126/science.1085953 [DOI] [PubMed] [Google Scholar]
- McBride, C. E., & Machamer, C. E. (2010). Palmitoylation of SARS-CoVs protein is necessary for partitioning into detergent-resistant membranes and cell-cell fusion but not interaction with M protein. Virology, 405(1), 139–148. 10.1016/j.virol.2010.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz, A., Shahid, F., Butt, T. T., Awan, F. M., Ali, A., & Malik, A. (2020). Designing multi-epitope vaccines to combat emerging coronavirus disease 2019 (COVID-19) by employing immuno-informatics approach. Frontiers in Immunology, 11(1663). 10.3389/fimmu.2020.01663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman, B. W., Adair, B. D., Yoshioka, C., Quispe, J. D., Orca, G., Kuhn, P., Milligan, R. A., Yeager, M., & Buchmeier, M. J. (2006). Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. Journal of Virology, 80(16), 7918–7928. 10.1128/JVI.00645-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarenko, J., Bui, H.-H., Li, W., Fusseder, N., Bourne, P. E., Sette, A., & Peters, B. (2008). ElliPro: A new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics, 9, 514. 10.1186/1471-2105-9-514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapin, N., Lund, O., Bernaschi, M., & Castiglione, F. (2010). Computational immunology meets bioinformatics: The use of prediction tools for molecular binding in the simulation of the immune system. PLoS One, 5(4), e9862. 10.1371/journal.pone.0009862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, A., Kucukural, A., & Zhang, Y. (2010). I-TASSER: A unified platform for automated protein structure and function prediction. Nature Protocols, 5(4), 725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samad, A., Ahammad, F., Nain, Z., Alam, R., Imon, R. R., Hasan, M., & Rahman, M. S. (2020). Designing a multi-epitope vaccine against SARS-CoV-2: An immunoinformatics approach. Journal of Biomolecular Structure and Dynamics, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaik, N. A. (2019). Essentials of bioinformatics, volume I: Understanding bioinformatics: genes to proteins. Springer International Publishing Imprint. [Google Scholar]
- Simmons, G., Reeves, J. D., Rennekamp, A. J., Amberg, S. M., Piefer, A. J., & Bates, P. (2004). Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proceedings of the National Academy of Sciences of the United States of America, 101(12), 4240–4245. 10.1073/pnas.0306446101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorp, E. B., Boscarino, J. A., Logan, H. L., Goletz, J. T., & Gallagher, T. M. (2006). Palmitoylations on murine coronavirus spike proteins are essential for virion assembly and infectivity. Journal of Virology, 80(3), 1280–1289. 10.1128/JVI.80.3.1280-1289.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E., & Berendsen, H. J. C. (2005). GROMACS: Fast, flexible, and free. Journal of Computational Chemistry, 26(16), 1701–1718. 10.1002/jcc.20291 [DOI] [PubMed] [Google Scholar]
- Vijgen, L., Keyaerts, E., Moës, E., Thoelen, I., Wollants, E., Lemey, P., Vandamme, A.-M., & Van Ranst, M. (2005). Complete genomic sequence of human coronavirus OC43: Molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. Journal of Virology, 79(3), 1595–1604. 10.1128/JVI.79.3.1595-1604.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne, G. (1984). Analysis of the distribution of charged residues in the N-terminal region of signal sequences: Implications for protein export in prokaryotic and eukaryotic cells. The Embo Journal, 3(10), 2315–2318. 10.1002/j.1460-2075.1984.tb02132.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waleed, M. S. (2020). Understanding the mosaic of COVID-19: A review of the ongoing crisis. Cureus, 12(3), e7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P., Sidney, J., Kim, Y., Sette, A., Lund, O., Nielsen, M., & Peters, B. (2010). Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics, 11(1), 568. 10.1186/1471-2105-11-568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Zhao, F., Peng, J., & Xu, J. (2011). Protein 8-class secondary structure prediction using conditional neural fields. Proteomics, 11(19), 3786–3792. 10.1002/pmic.201100196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, T. K., Jordan, R., Lo, M. K., Ray, A. S., Mackman, R. L., Soloveva, V., Siegel, D., Perron, M., Bannister, R., Hui, H. C., Larson, N., Strickley, R., Wells, J., Stuthman, K. S., Van Tongeren, S. A., Garza, N. L., Donnelly, G., Shurtleff, A. C., Retterer, C. J., … Bavari, S. (2016). Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature, 531(7594), 381–385. 10.1038/nature17180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, S. K., Li, W., Moore, M. J., Choe, H., & Farzan, M. (2004). A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. The Journal of Biological Chemistry, 279(5), 3197–3201. 10.1074/jbc.C300520200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, P. C. Y., Lau, S. K. P., Chu, C.-M., Chan, K.-H., Tsoi, H.-W., Huang, Y., Wong, B. H. L., Poon, R. W. S., Cai, J. J., Luk, W.-K., Poon, L. L. M., Wong, S. S. Y., Guan, Y., Peiris, J. S. M., & Yuen, K.-Y. (2005). Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. Journal of Virology, 79(2), 884–895. 10.1128/JVI.79.2.884-895.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, X., Chakraborti, S., Dimitrov, A. S., Gramatikoff, K., & Dimitrov, D. S. (2003). The SARS-CoV S glycoprotein: Expression and functional characterization. Biochemical and Biophysical Research Communications, 312(4), 1159–1164. 10.1016/j.bbrc.2003.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Y., Tao, H., He, J., & Huang, S.-Y. (2020). The HDOCK server for integrated protein-protein docking. Nature Protocols, 15(5), 1829–1852. 10.1038/s41596-020-0312-x [DOI] [PubMed] [Google Scholar]
- Yi, Y., Lagniton, P. N. P., Ye, S., Li, E., & Xu, R.-H. (2020). COVID-19: What has been learned and to be learned about the novel coronavirus disease. International Journal of Biological Sciences, 16(10), 1753–1766. 10.7150/ijbs.45134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H.-Q., Ning, L.-W., Zhang, H.-X., & Guo, F.-B. (2014). Analysis of the relationship between genomic GC Content and patterns of base usage, codon usage and amino acid usage in prokaryotes: Similar GC content adopts similar compositional frequencies regardless of the phylogenetic lineages. PLoS One, 9(9), e107319. 10.1371/journal.pone.0107319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla, A., Chan, J. F. W., Azhar, E. I., Hui, D. S. C., & Yuen, K.-Y. (2016). Coronaviruses-drug discovery and therapeutic options. Nature Reviews. Drug Discovery, 15(5), 327–347. 10.1038/nrd.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.