Abstract
Novel Coronavirus or SARS-CoV-2 has received worldwide attention due to the COVID-19 pandemic, which originated in Wuhan, China leading to thousands of deaths to date. The SARS-CoV-2 Spike glycoprotein protein is one of the main focus of COVID-19 related research as it is a structural protein that facilitates its attachment, entry, and infection to the host cells. We have focused our work on mutations in two of the several functional domains in the virus spike glycoprotein, namely, receptor-binding domain (RBD) and heptad repeat 1 (HR1) domain. These domains are majorly responsible for the stability of spike glycoprotein and play a key role in the host cell attachment and infection. In our study, several mutations like R408I, L455Y, F486L, Q493N, Q498Y, N501T of RBD (319-591), and A930V, D936Y of HR1 (912-984) have been studied to examine its role on the spike glycoprotein native structure. Comparisons of MD simulations in the WT and mutants revealed a significant de-stabilization effect of the mutations on RBD and HR1 domains. We have investigated the impact of mapped mutations on the stability of the spike glycoprotein, before binding to the receptor, which may be consequential to its binding properties to the receptor and other ligands.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, SARS-CoV-2 spike glycoprotein, RBD and HR1 domain, deleterious mutations, MD simulations
Introduction
COVID-19 is majorly responsible for the fatal respiratory disorder in severely infected patients that has become one of the concerns globally, though, the typical symptoms range from mild cold and fever (Coutard et al., 2020; Tang et al., 2020). The pathogenic novel coronavirus has now affected more and more individuals with the rise in the death toll too. The pandemic threat to humans is getting more severe with the sky-scraping changes in the number of deaths every minute (Giurgea et al., 2020). Worldwide efforts are on to curtail the rapid spread of the virus, by developing effective strategies for patient management, effective novel/repurposed drugs and vaccine (Cascella et al., 2020; Giurgea et al., 2020).
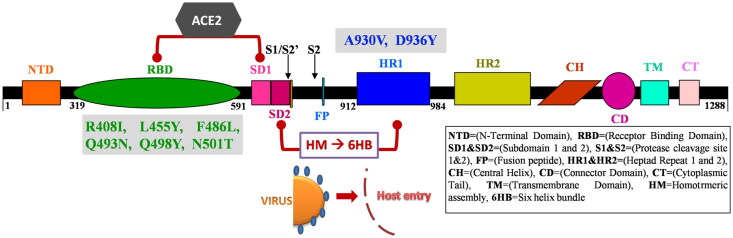
The SARS-CoV-2 spike glycoprotein has grabbed the attention of the worldwide research groups for its efficient function to enable the virus to attach and infect host cells. The protein, with highly conserved domains and motifs, is 1288 amino acid residue long and belongs to the class-I viral proteins, which contributes to receptor-binding and pathogenesis (Choudhury & Mukherjee, 2020). The SARS-CoV-2 spike glycoprotein has differential forms of homotrimers ejecting out from the viral surface (Wrapp et al., 2020). The Cryogenic Electron Microscopy (Cryo-EM) structure of the spike glycoprotein shows that it contains two S1 and S2 subunits, where the Receptor-Binding Domain (RBD) covers the segment from 319-591 in the S1 subunit containing a loop region from residues 424-494 which directly come in contact with Angiotensin-Converting Enzyme 2 (ACE2) (Lan et al., 2020). After the binding of the RBD of the S1 subunit with ACE2 on the host cell, Heptad Repeat domains (HR1 and HR2) located in the S2 subunit interact to create a proximity contact between the virus and host entry. This S2 subunit forms a fusion peptide in this process shaping a homotrimeric assembly with the HR1 domain to structure three hydrophobic grooves on the surface (Figure 1) (Lokman et al., 2020). The furin cleavage site located between the S1 and S2 subunits preactivates the SARS-CoV-2 virus entry into the host cell (Andersen et al., 2020; Shang, Wan et al., 2020). The assembly of both S1 and S2 subunits, with the involvement of RBD and HR1 domains, along with various other domains on the spike glycoprotein helps in forming a Six Helix Bundle (6HB) on the virus which facilitates the virus entry into the host cells (Lokman et al., 2020).
Figure 1.
Schematic representation of SARS-CoV-2 spike glycoprotein functional domains/motifs with the representation of ACE2 binding on RBD (Green), 6HB formation through HR1 domain (Blue) and mutation sites in the process of virus-host entry.
The recent literature mining has explored the RBD residues binding to the ACE2 receptor (Andersen et al., 2020; Lokman et al., 2020). Shang, Wan, et al., (2020) highlighted the importance of the ACE2 binding with RBD of SARS-CoV2 in the discovery of feasible drugs and vaccines that can block the virus-host entry (Shang et al., 2020a). The SARS-CoV-2 RBD domain residues like Lys417, Leu455, Phe486, Gln493, Gln498, and Asn501 (hydrogen bond) bind to ACE2 receptor with the residues Asp30, Gln24, Lys31, Asp38, and Try41 revealing the significance of RBD as a functional component (Brielle et al., 2020; Shang et al., 2020b). ACE2 binds with the SARS-CoV-2 RBD region forming hydrogen interactions which in turn induces the S1 subunit. Soon after, the HR1 domain of the S2 subunit forms the membrane fusion and initiates the step of viral host entry (Brielle et al., 2020; Lokman et al., 2020; Shang et al., 2020a). Hence, with this plinth of background, RBD and HR1 domain of SARS-CoV-2 spike glycoprotein were selected based on its indispensable role in the host entry of the virion and subsequent pathogenicity progression (Figure 1).
There is a vast amount of genome sequencing data and extensive literature evidence which indicate that the selected RBD, as well as HR1 residues on SARS-CoV-2 spike glycoprotein, are majorly having strong binding with human ACE2 residues and also involved in the disease progression (Supplemental Table S1 & S2). To analyze and make use of this data, computational approaches are more powerful, inexpensive, time effective and involves minimal man-power. The single amino acid mutation is directly proportional to the functional contacts of protein/receptor within the complex diseases and also known for its quick drug response (Schreiner et al., 2011). Many cases of studies have been reported for the successful application of mutation associated strategy to understand the individual drug response, phenotypic variability at genetic state in personalized medicine, as this area of study utilizes sequence, structural, physicochemical and stereo-chemical properties of amino acids (Pires et al., 2017; Schreiner et al., 2011).
Jia et al. reported various mutations on spike glycoprotein and also highlighted the importance of R408I mutation responsible for lower ACE2 binding which revealed a higher risk of COVID-19 pandemic (Jia et al., 2020). Ou et al. implemented Molecular Dynamics (MD) simulation studies of 10 ns for each identified mutations to understand the de-stabilizing role on spike glycoprotein (Ou et al., 2020). The MD simulation study was useful to understand the residue contacts within the spike glycoprotein which can form or break a bond with a change in single nanosecond (Perilla et al., 2015). We have performed 100 ns MD simulations for the wild type (WT) Spike glycoprotein, its RBD and HR1 domain mutants to understand the fundamental atomic movements and conformational changes in the mutated structures. In this work, the three-dimensional (3D) prefusion structure of the SARS-CoV-2 spike glycoprotein (PDB ID: 6VSB) RBD region from 319 to 591 as well as the HR1 segment from 912 to 984 was trimmed to constitute WT (Figure 1). The important RBD mutations like R408I, L455Y, F486L, Q493N, Q498Y, N501T and the mutations A930V, D936Y mapped on HR1 domain were derived due to its significant role in protein de-stabilization (Brielle et al., 2020; Lokman et al., 2020; Perilla et al., 2015; Shang et al., 2020b, 2020). The MD trajectories of WT and mutant structures were compared using Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), Radius of gyration (Rg), Solvent Accessible Surface Area (SASA) and Free Energy Landscape (FEL) analysis (Amir et al., 2019; Kanipakam et al., 2020). The mutations were analyzed using various computational algorithms and MD simulations enabled to predict the impact of the deleterious mutations R408I, L455Y, F486L, Q493N, Q498Y, N501T of RBD and HR1 associated A930V, D936Y mutations in the prefusion SARS-CoV-2 spike glycoprotein (Adzhubei et al., 2013; Rodrigues et al., 2018; Worth et al., 2011). Thus, our findings provide useful insights for the research studies aimed at targeting the SARS-CoV-2 spike protein and the ACE2 receptor binding interface residues for the development of novel inhibitors against COVID-19.
Material and methods
Generation of mutant structures
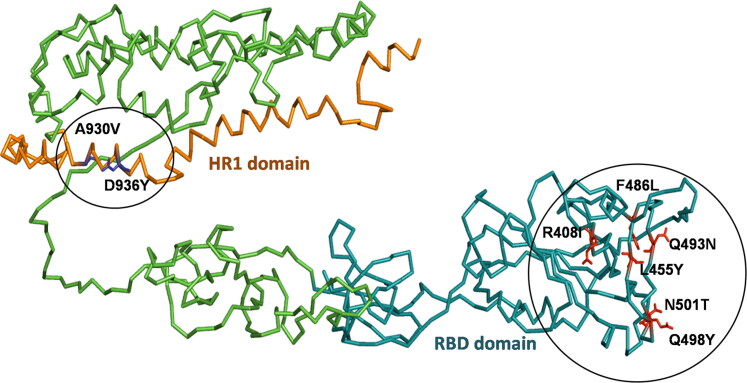
The 3D structure of human SARS-CoV-2 spike glycoprotein was retrieved from PDB (ID: 6VSB) with an amino acid length coverage of 1288 and resolution 3.46Ǻ (Wrapp et al., 2020). The R408I, L455Y, F486L, Q493N, Q498Y, N501T mutations on the RBD region and A930V, D936Y mutations on the HR1 domain were mutated at respective positions (Figure 2). The shortlisted mutations were introduced into the spike glycoprotein structure using SwissPDB viewer and hence ten structures were generated (Guex & Peitsch, 1997; Johansson et al., 2012; Schwede et al., 2003).
Figure 2.
Illustration of RBD (Aqua Blue) and HR1 (Orange) domains annotated on spike glycoprotein in wire model, generated using PyMOL.
Prediction of the effect of mutations
The prediction methods SIFT, PolyPhen, I-mutant 2.0, PROVEAN and PhD-SNP (Sim et al., 2012), were employed to determine the individual role of RBD and HR1 mapped mutations on the function of SARS-CoV-2 spike glycoprotein. Further, the destabilizing effects of mutations at the substituted positions were also confirmed by predicted changes in folding Gibbs free energy (ΔΔG) by DUET, mCSM, EnCoM, DynaMut and SDM web servers which provide the threshold scores effectively (Amir et al., 2019, 2020; Pires et al., 2014). From these examinations, de-stabilizing, damaging and deleterious RBD and HR1 mutations of the spike glycoprotein receptor were identified.
Molecular dynamics simulation
MD simulations were carried out using the GROMACS 5.18.3 software package (Hess et al., 2008; Pronk et al., 2013). The crystal structure of SARS-CoV-2 spike glycoprotein was collected, RBD and HR1 domains were modeled by the SWISS-MODEL online server (Schwede et al., 2003) to accurate the missing residues in the native PDB structure. The WT of both the domains and all the mutations topology parameter files were created by the CHARMM27 force field. The intermolecular (non-bonded) potential, represented as the sum of Lennard–Jones (LJ) force and pairwise Coulomb interaction and the long-range electrostatic force, were determined by particle mesh Ewald approach. The velocity Verlet algorithm was applied for the numerical integrations and the initial atomic velocities were generated with a Maxwellian distribution at the given absolute temperature. Then the system immersed with the SPC/E water model, and protein was placed at the center of the cubic grid box (of dimension 1.0 nm3) (Zielkiewicz, 2005). The neutralization was performed and to make concentration to 0.15 M. The neutralized system was then subjected to energy minimization using the Steepest Descent (SD) and Conjugate Gradient (CG) algorithms utilizing a convergence criterion of 0.005 kcal mol−1 Å−1. The two-standard equilibration phase was carried out separately NVT (atom, volume, temperature) and NPT (atom, pressure-temperature) ensemble conditions such as constant volume and constant pressure for each complex similar time scale. We applied the Berendsen thermostat and Parinello–Rahman barostat to maintain the temperature and the pressure of the system, respectively. The system was maintained constant at 1 bar and 300 K, with a coupling time of τp = 2 ps, and τT = .1 ps, respectively (Figure 3).
Figure 3.
Schematic representation of the in silico workflow followed to study the impact of mutations on the stability of the spike glycoprotein RBD and HR1 domains.
The Periodic Boundary Condition (PBC) used for integrating the equation of motion by applying the leap-frog algorithm with a 2 fs time step. Finally, to make the system ready for production the fixing of constraints is achieved with the relaxation of the grid box with water along with the counterions. In the current study, all the simulations, including RBD WT and its six mutant structures; HR1 WT and its two mutant structures, were subjected to 100 ns simulations, each (Ahamad et al., 2018, 2019).
Principal component analysis (PCA)
PCA of the MD trajectories or Essential Dynamics (ED) was performed to reduce the complexity of the simulation-coordinates to understand the most significant motions. The eigenvector and eigenvalues obtained by diagonalizing covariance matrix and the atomic movements of two Principal Components (PC1 and PC2) were examined by the ED method (Amir et al., 2019, 2020). The movement and the dynamic motions of each atom are marked on both WT (RBD & HR1) and mapped mutant structures through total simulation run time and the trajectory subspaces which were further validated by Cartesian trajectory co-ordinates by projecting the crucial eigenvectors.
Analysis of the molecular motions
The molecular motions in the systems- WT and mutant structures are examined with MD simulation, and the coordinates were saved at each interval from the obtained trajectories. The trajectory files were analyzed further to spot the positional deviations, relative fluctuations, direction of magnitude, change in atomic motions and hydrogen bond formations (Ahamad et al., 2018, 2019; Kanipakam et al., 2020). The parameters were calculated for both WT (RBD & HR1) and mutant structures with different modules to identify the contributions of each atom in the protein that influences the structural stability and function of the prefusion SARS-CoV-2 spike glycoprotein.
Free energy landscape (FEL) and frustration analysis
The conformational changes that occur at every time step of the simulation and varies from WT in the mutant structures were investigated with FEL analysis (Figure 3). With this analysis the protein that attains a specific position can be marked which consumes some amount of energy on the energy landscapes (Sang et al., 2017). The FEL study was implemented on both the systems (WT-RBD/HR1 and mapped mutations) through eigenvectors of PC1 and PC2 lane. The energetically favored and the intra-molecular contacts of each residue for both WT (RBD & HR1) and mutant structures were determined with frustratometer analysis (Parra et al., 2016). The single and configuration index residues were set between the X and Y axis with a threshold Z-scores. The difference in the Z-score values with <-0.78 was grouped as de-stable, >0.78 as stable or minimal frustrating and in between the scores were marked as neutral. The WT (RBD & HR1) and mutant structures were subjected to the frustration analysis to determine the spike glycoprotein destabilizing and the stabilizing energies.
Results and discussion
In silico screening of the mutations
The functional effects of R408I, L455Y, F486L, Q493N, Q498Y, N501T mutations in the RBD domain, and A930V, D936Y mutations in the HR1 domain were predicted using SIFT, PolyPhen, I-mutant 2.0, PROVEAN, PhD SNP online servers. SIFT analysis provided the tolerance index score of all the mutations between 0 to 1 revealing its deleterious effect on the amino acid substitution at the respective positions on the protein which was ranked based on the aligned sequences. PolyPhen predicted all the mutations to be deleterious with almost unity score in all the mutations (Table 1). I-mutant 2.0 predicted all the mutations were altering the stabilization, acquired from an amino acid change with increased or decreased stability. The PROVEAN scores also confirmed the mutations to confer a damaging effect on the native structure. PhD-SNP method also projected that the selected mutations fall under disease-related variants based on the local sequence environment. The screening results indicate the deleterious and pathogenic effects of all the mutations on the 3D structure of spike glycoprotein by altering its stability and function (Table 1).
Table 1.
Predicted effect of protein stability in the presence of amino acid mutations in the spike glycoprotein- RBD and HR1 domains.
| S. No | Mutants | SIFT Score | PROVEAN Score | PolyPhen Score | PhD SNP Score | i-mutant (DDG) | DUET (ΔΔG) | mCSM (ΔΔG) | EnCOM (ΔΔG) | DynaMut (ΔΔG) | SDM (ΔΔG) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RBD associated six mutants | |||||||||||
| 1 | R408I | 0.38 Deleterious |
0.225 Damaging |
0.999 Destabilizing |
8 Diseased |
−0.20 kcal/mol |
−0.621 kcal/mol |
−0.562 kcal/mol |
−4.327 kcal/mol |
−1.857 Destabilizing |
−0.160 Destabilizing |
| 2 | L455Y | 1.00 Deleterious |
1.247 Damaging |
0.900 Destabilizing |
7 Diseased |
−1.42 kcal/mol |
−1.098 kcal/mol |
−0.789 kcal/mol |
−4.109 kcal/mol |
−1.662 Destabilizing |
−0.973 Destabilizing |
| 3 | F486L | 0.66 Deleterious |
−0.017 Damaging |
0.991 Destabilizing |
8 Diseased |
−0.31 kcal/mol |
−1.824 kcal/mol |
−0.336 kcal/mol |
−4.842 kcal/mol |
−1.320 Destabilizing |
−0.864 Destabilizing |
| 4 | Q493N | 0.67 Deleterious |
0.631 Damaging |
0.999 Destabilizing |
7 Diseased |
−0.82 kcal/mol |
−0.107 kcal/mol |
−0.556 kcal/mol |
−4.989 kcal/mol |
−1.694 Destabilizing |
−0.250 Destabilizing |
| 5 | Q498Y | 1.00 Deleterious |
−1.835 Damaging |
0.990 Destabilizing |
8 Diseased |
−0.02 kcal/mol |
−0.224 kcal/mol |
−0.064 kcal/mol |
−4.189 kcal/mol |
−1.749 Destabilizing |
−0.910 Destabilizing |
| 6 | N501T | 1.00 Deleterious |
1.199 Damaging |
0.991 Destabilizing |
8 Diseased |
−0.86 kcal/mol |
−1.954 kcal/mol |
−0.826 kcal/mol |
4.982 kcal/mol |
−1.210 Destabilizing |
−0.986 Destabilizing |
| HR1 associated two mutants | |||||||||||
| 1 | A930V | 0.46 Deleterious |
−3.725 Damaging |
0.999 Destabilizing |
4 Diseased |
−2.64 kcal/mol |
−0.343 kcal/mol |
−0.380 kcal/mol |
−4.004 kcal/mol |
−1.321 Destabilizing |
−0.343 Destabilizing |
| 2 | D936Y | 1.00 Deleterious |
−2.569 Damaging |
0.988 Destabilizing |
5 Diseased |
−0.17 kcal/mol |
−0.420 kcal/mol |
−0.499 kcal/mol |
−4.648 kcal/mol |
−1.299 Destabilizing |
−0.130 Destabilizing |
SIFT; sorting intolerant to tolerant, PhD-SNP; predictor of human deleterious single nucleotide polymorphism, PROVEAN; Protein Variation Effect Analyzer; SDM; site directed mutator..
Effect of Gibbs free energy (ΔΔG)
The effect on stability due to changes in R408, L455, F486, Q493, Q498, N501 to 408I, 455Y, 486L, 493N, 498Y, 501T, respectively, in RBD; and A930, D936 of HR1 to 930 V, 936Y, respectively, were further evaluated using Gibbs free energy calculation (ΔΔG) with DUET, mCSM, EnCOM, DynaMut and SDM web servers (Frappier et al., 2015; Pandurangan et al., 2017; Rodrigues et al., 2018). The results depicted that the mutations render de-stabilizing effects (Supplemental Figure S1 1 A-C). The employed dynamic programs revealed the deleterious impact of R408I, L455Y, F486L, Q493N, Q498Y, N501T mutations in the RBD and A930V, D936Y mutations in the HR1 domain on the stability of the SARS-CoV-2 spike glycoprotein native structure (Table 1).
Protein deviation and compactness analysis
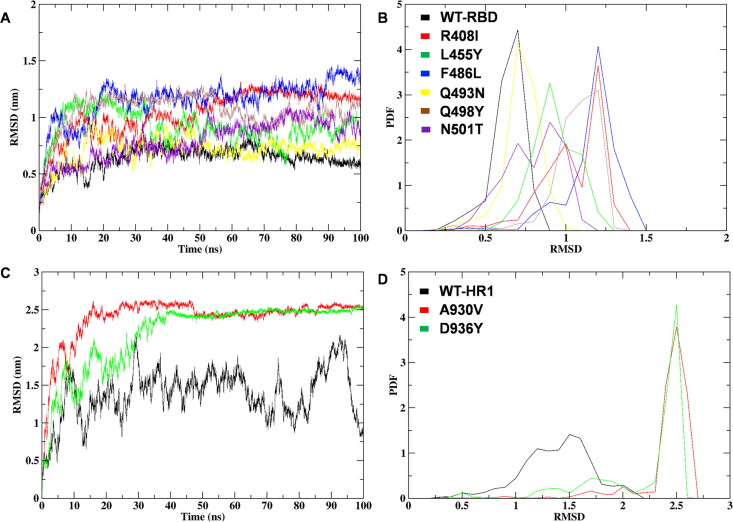
The MD trajectories of WT of RBD, HR1 and the mutant structures were analyzed by RMSD calculations. The MD simulation of the WT of the RBD region noticeably displays a steady RMSD of ∼0.5 nm, throughout the simulation, whereas L455Y shows a fluctuation of ∼0.8 nm, followed by Q493N, Q498Y, N501T structures with a range of ∼0.7 nm, respectively. Interestingly, the maximum drift of fluctuation was noticed in R408I, F486L with an RMSD range of ∼1.0 nm (Figure 4A). The results were further investigated for the Probability Distribution Function (PDF) for the obtained RMSD averages of RBD WT and mutants. The analysis revealed that the probability distribution of WT was within the threshold of 0.6 nm whereas the R408I, L455Y, F486L, Q493N, Q498Y, N501T mutant structures revealed maximum alterations of 1.0 nm, 0.9 nm, 1.1 nm, 0.7 nm, 0.7 nm, 0.7 nm, respectively (Figure 4B). The analysis of the RMSDs of the HR1 domain mutants revealed a minimum constant range of ∼1.3 nm for WT, whereas the highest fluctuations were observed for the structures with mutations A930V, D936Y with RMSD in the range of ∼2.0 nm (Figure 4C). The PDF investigations revealed WT with 1.5 nm and mutants exhibited a high drift of fluctuations for A930V, D936Y with 2.7 and 2.5 nm of average PDF-RMSD ranges (Figure 4D) and (Supplemental Table S3). The deviation results of RBD and HR1 domains marked major differences in the spike glycoprotein C-alpha (Cα) atom conformational change leading to protein de-stabilization effects due to the presence of mutations. The differences in the range of deviations reflect the significant role of amino acid substitution on SARS-CoV-2 spike glycoprotein native 3D structure on the Cα atom of protein. Results also revealed that there is a maximum effect on the flexibility of spike glycoprotein structure due to the presence of positioned mutations on RBD and HR1 domains.
Figure 4.
MD simulation RMSD analysis of mutated SARS-CoV-2 spike glycoprotein RBD and HR1 domains (A) Comparison plot of RMSD between WT and mutant structures of RBD (B) Probability distribution function of RMSD of WT and mutants of RBD showing the altered ranges on protein stability (C) RMSD comparison of HR1 domain WT and mutants (D) PDF fluctuation plot of HR1 domain WT and mapped mutants.
Root-mean square fluctuations
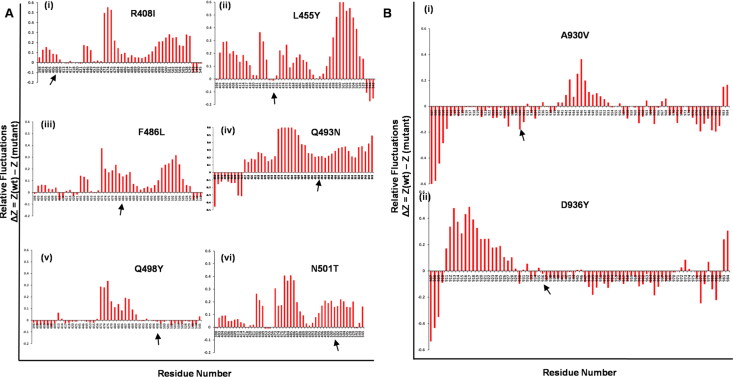
The overall flexibility of the RBD-WT and R408I, L455Y, F486L, Q493N, Q498Y, N501T mutant structures, as well as HR1-WT and A930V, D936Y structures were analyzed with Cα-RMSF relative fluctuations (Supplemental Figure S2A-B). The negative drift of the graph indicated the increased movement of atoms on both RBD and HR1 domains by mutations are predicted to be the cause of instability of the native receptor in comparison to WT. RBD mutant R408I revealed Q414, Y421, F543, N544, G545 residues to have high negative fluctuations, L455Y mutant demonstrated higher fluctuations at position D398, Y453, F456, F543, N544, G545, F486L mutant revealed maximum fluctuations on residues D398, G413, Q414, D420, Y421, F543, N544, G545, and Q493N revealed high fluctuations at positions D398, R403-Q409, G413, and Q414 respectively (Figure 5A(i–iv)). The mutation Q498Y revealed highest fluctuations on the maximum number of residues positioned at D398, R403-Q409, K417-Y421, Y449, Y453, L455, L492, Q493, G496, Q498, P499, T500, G502-Y505, K528, K529, F543, N544, followed by N501T with fluctuations at D398, Y453, L455, and F543 respectively (Figure 5A(v–vi)). Spotting HR1 domain, mutant A930V was marked with projected higher fluctuations on residues I907-I931, K933, I934, Q935, L938, S939, Q957, A958, N960, T961, L962, K964, Q965, L966, N969-A972, and S974-S982 followed by, mutant D936Y showed higher relative alterations at positions I907-G910, A930, K933, I934, Q935-A944, K947-D970, V976-N978, I980-S982, respectively (Figure 5B(i, ii)). Intriguingly, we observed that the fluctuations were lower at the mutated residues. The internal motion of each residue was averaged to the relative fluctuation to get the comparative graph between mutations and WT of both the domains located on spike glycoprotein. Thus, it was observed that mutation brings about higher fluctuations mainly in the RBD region around 400-544 and HR1 domain around 930-940, where the mapped mutations are rolled. The positioned mutations like R408, L455, F486, Q493, Q498, N501 mutations of RBD that are associated in the binding of ACE2 receptor as well as A930, D936 mutations of HR1 interestingly display positive value, implying that mutation mainly imparts rigidity of spike glycoprotein leading to the greatest degree of flexibility loss to the WT structure.
Figure 5.
Root Mean Square Fluctuations (RMSF) of both the spike glycoprotein domains showing the peak drift in the mutant structures at residue level A(i-vi). The six RBD domain mapped mutations B(i-ii). The two HR1 mutations (position of mutations are indicated with black arrows).
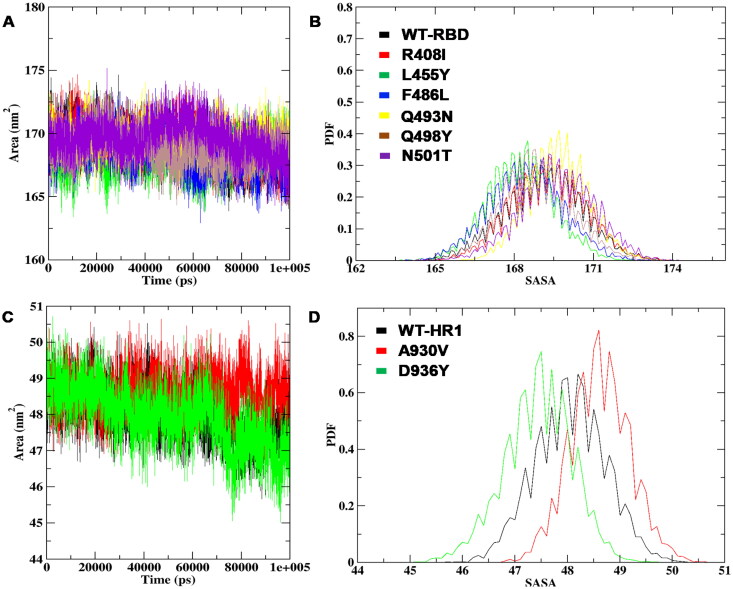
Solvent-accessible surface area and Radius of gyration
The SASA and the measure of the compactness of structures were monitored for WT and the mutant structures of both the RBD and HR1 domains throughout the MD simulation. The SASA analysis revealed that the R408I, Q493N, Q498Y, and N501T had a higher range of SASA values with a range of 169nm2, followed by L455Y, F486L with 168nm2, whereas WT showed a minimum range of SASA (Figure 6A). The PDF of the obtained results revealed maximum drifts for the mutants R408I, L455Y, F486L, Q493N, Q498Y, N501T with a range of 169.24nm2, 168.23nm2, 168.42nm2, 169.65nm2, 169.61nm2, 169.62nm2, where are WT was shown with 169.10nm2 respectively (Figure 6B). Subsequently, the SASA of mutants A930V and D936Y of HR1 domain projected maximum fluctuations with 49nm2 range and WT was with 48nm2, respectively (Figure 6C). The decrease in SASA was noticed on mutation D936Y with a PDF-SASA average of 47nm2 and sharp alteration peaks were spotted for A930V with 49nm2 range,in contrast, 48nm2 range was noticed on WT for HR1 mutations (Figure 6D) and (Supplemental Table S3). The alterations in the SASA values, when compared to WT, justified high loss of hydrophobic contacts on both RBD and HR1 domains with the presence of mutants to destruct secondary structural elements on native SARS-CoV-2 protein. Interestingly the mutations on the RBD region like L455Y, F486L, Q493N, and N501T that forms binding with ACE2 receptor were observed to be with the highest deviations. Thereby, it was correlated that the R408I, L455Y, F486L, Q493N, Q498Y, N501T mutations on the RBD region and A930V, D936Y mutations are in turn responsible for the de-stabilizing the native spike glycoprotein further probably halting the process of forming a 6HB on the virus to host entry.
Figure 6.
SASA analysis of RBD and HR1 WT structures in the presence of mutations (A) SASA plot of RBD region with mapped mutations (B) The consistency probability energy plot of RBD WT and mutations (C) SASA fluctuations per residue variation for HR1 domain WT and mutations (D) The PDF plot representing the SASA average values on WT and mutants of HR1 domain at total simulation time.
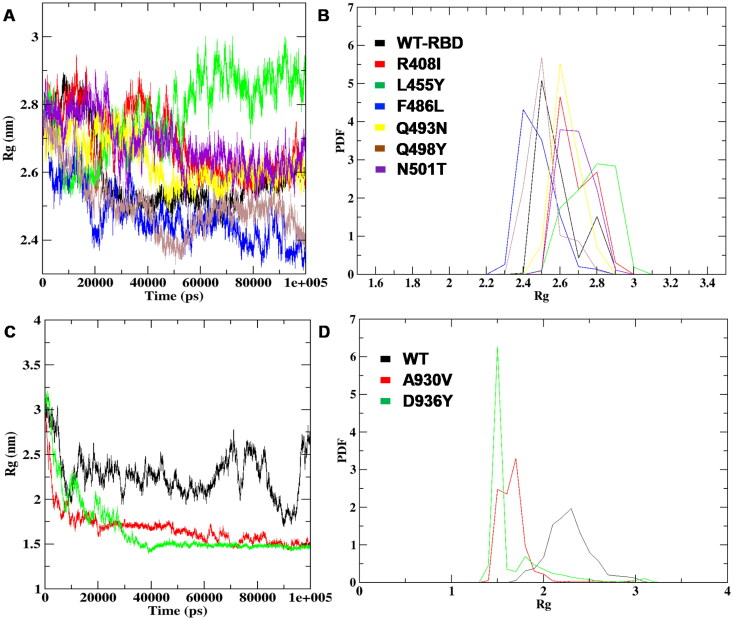
To understand the RMS-distance of each atom from the center, the radius of gyration (Rg) analysis was implemented on ten mutant structures along with WT’s. It was noticed that the RBD mapped mutations R408I, L455Y, F486L, Q493N, Q498Y, and N501T showed the average highest Rg value of ∼2.6 nm and WT was with 2.5 nm (Figure 7A). The study was further investigated to note the PDF between the WT and mutants revealed there was an unsteady drift in the graph with average Rg of noticed on R408I with 2.68 nm, L455Y with 2.77 nm, F486L with 2.47 nm, Q493N with 2.63 nm, Q498Y with 2.68 nm and N501T with 2.68 nm, whereas the difference in value was noticed on WT with 2.5 nm (Figure 7B). The comparative analysis of HR1 WT and the mutants showed A930V and D936Y presented the least compactness with the highest deviation score of ∼1.0 nm noticed, whereas ∼2.0 nm of Rg was marked on WT in all the time scales of generated trajectories, respectively (Figure 7C). The PDF analysis of A930V and D936Y mutations further confirmed the loss of gyration between the atoms in the presence of A930V with an Rg average of 1.67 nm, D936Y with 1.65 nm revealing decreased peaks when compared to WT with 2.27 nm of Rg average (Figure 7D) and (Supplemental Table S3). The analysis of SASA and Rg data suggested the compactness, area of stability was consistent for both WT structures and R408I, L455Y, F486L, Q493N, Q498Y, N501T mutations on RBD region and A930V, D936Y mutations of HR1 altered maximally towards the end of simulations leading to structural destabilization, loss of shape and folding on spike glycoprotein native structure.
Figure 7.
Rg plots of RBD and HR1 domain WT and mutants structures (A) The radius of gyration consistency plot mapped to compare RBD-WT and mutants (B) RBD WT and mutant Probability distribution function of Rg (C) Radius of gyration plot annotated on HR1 domain WT and mutant structures (D) PDF plot representation between the HR1 domain WT and mutant structures.
Intra-molecular hydrogen bond analysis
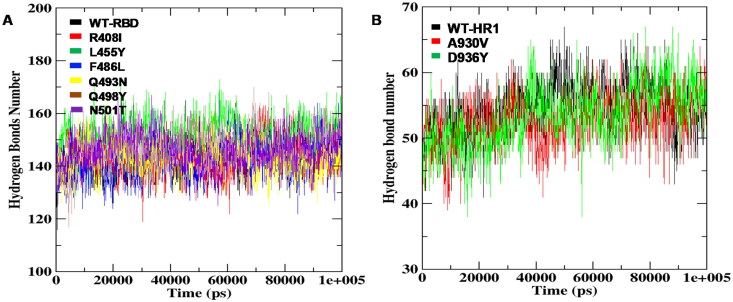
The change in hydrogen bond numbers formed in the structures of RBD and HR1 WT was compared with the respective mutants during the simulation run. The analysis projected that RBD-WT revealed 166.41 hydrogen bonds which were consistent from 0 ns to 100 ns. The intramolecular hydrogen bonds found low numbering on mutation structures like F486L and Q498Y with 142.50 and 146.87, whereas, the average number of hydrogen bonds were noticed on RBD mutants R408I, L455Y, Q493N, and N501T with 166.55, 166.71, 167.01, 167.28, respectively (Figure 8A). A lower number of hydrogen bonds were observed on HR1 mutations A930V, D936Y with 55.98 and 59.12, in contrast, WT-HR1 formed 60.70 hydrogen bonds during the run time (Figure 8B). Remarkably, it was also observed that the RBD residue N501 involved in hydrogen bond formation with ACE2 has shown alterations in forming the intra-molecular bond number in the presence of mutation N501T revealing the damaging effect on the RBD loop lying from 319-591 on spike glycoprotein (Supplemental Table S4). From the overall analysis, it was interpreted that the loss of rigidity on native SARS-CoV-2 spike glycoprotein structure was due to the high differences in hydrogen bond formation which in turn creating high negative pressure on RBD region and HR1 domain, disturbing the formation of 6HB, accountable for infection to arise.
Figure 8.
Intramolecular hydrogen bond monitoring of RBD and HR1 domains (WT and mutant structures) (A) Hydrogen bond monitoring between RBD-WT and mutant structures for total MD simulation run time (B) HR1 domain WT and mutant structures forming hydrogen bonds throughout the total run time of 100 ns each.
Dynamic behavior analysis
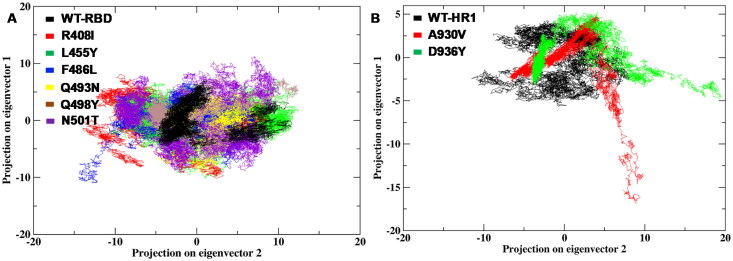
The essential subspace gives the maximum protein dynamics that can be noted with eigenvectors which are largely associated with the eigenvalues. The dynamic behavior of RBD and HR1 domain WT was compared to the mutant structures with the above-mentioned parameter behind MD simulations. The results revealed that the clusters are well defined in the WT structures of the RBD and HR1 domain by covering the minimum region (Figure 9A and 9B). The mutants R408I, L455Y, F486L, Q493N, Q498Y, N501T mutations on RBD region and A930V, D936Y mutations of HR1 covered large region along the PC1 and PC2 lanes particularly compared to WT.
Figure 9.
The dynamic energy fluctuation plot between eigenvector 1 and vector 2 generated between both the systems (WT & mutants) of RBD and HR1 domain (A) Atomic number monitoring between eigenvector1 and vector2 of RBD region WT and mutant (B) The 2D projection plot between two eigenvectors for the conformational space of Cα-atom between HR1 WT and mutant structures.
Further, the eigenvectors vector1 and vector2 were plotted which gave the displacement of atomic fluctuation, which indicates the type of motion obtained between the WT and mutants of both RBD and HR1 domains of spike glycoprotein. The overall results suggested that R408I, L455Y, F486L, Q493N, Q498Y, N501T mutations on RBD region showed the highest fluctuation of ∼0.2 nm to ∼1.0 nm, whereas WT revealed highest drift of ∼0 to 0.2 nm throughout the MD simulations (Supplemental Figure S3A-B) and the A930V, D936Y mutations of HR1 domain showed unstable fluctuation of ∼0.2 nm to ∼1.5 nm, distinguished with WT range of ∼0.2 nm, respectively (Supplemental Figure S3C-D). The comparison of WT and mutant of RBD & HR1 domains showed unsteady fluctuations in the drift of eigenvalues which were marked against eigenvectors on both the domain systems. The results from the analysis of eigenvector suggested a distinguishable conformational change on RBD and HR1 domains leading to disturbance in the internal motion that was majorly responsible for spike glycoprotein un-stability.
The systems (RBD & HR1) WT and mutants were further examined with a diagonalized covariance matrix to understand the positional fluctuations on Cα-atoms to examine the atomic behavior of the correlated and anti-correlated motions. The analysis was illustrated with two intense color representations, red and blue. The small fluctuations between the atoms were marked with blue and the large fluctuations were denoted with red. The amplitude and the intensity of blue were magnified in WT structures of RBD and HR1 domains which were minimum with 0.8nm2 and 1.6nm2 leading to fluctuation consistency (Supplemental Figure S4A(i)–B(i)). The results were contrasting in the mutant structures with direction and the magnitude of motion altered drastically. The increase in the residue displacement was observed on RBD mapped mutations ranging from 1nm2 to 2nm2 and HR1 mutants with 1.8nm2 to 2nm2 respectively (Supplemental Figure S4A(ii–vii)-B(ii–vii)). The overall degradation of residue displacement was noticed due to the presence of R408I, L455Y, F486L, Q493N, Q498Y, N501T mutations on the RBD region and A930V, D936Y mutations on HR1 domains in turn de-stabilizing the SARS-CoV-2 spike glycoprotein crude structure.
FEL analysis
The conformational changes of spike glycoprotein specifically with respect to RBD and HR1 domains were observed for the FEL analysis that are directly proportional to its function that varies from several seconds. In this scenario, the deep investigation of dynamic conformational changes of WT of both domains and mutations involved in FEL was obtained with eigenvectors (Supplemental Figure S5A-5B). The global free energy minima (marked with KJ/mol) that are confined in the case of RBD and HR1 WT revealing the steady states, in contrast, the observations marked differences in the folding behavior on R408I, L455Y, F486L, Q493N, Q498Y, N501T mutations on RBD region (Supplemental Figure S5A(i–vii)) and A930V, D936Y mutations of HR1 domain (Supplemental Figure S5B(i–iii)). The overall analysis revealed significant alterations in the change of conformations on the mutants with high energy minima suggesting the substitutions of amino acids at mutant occurrence positions are leading to an unfolding pattern on the RBD and HR1 domains leading to SARS-CoV-2 spike glycoprotein stability and functional loss.
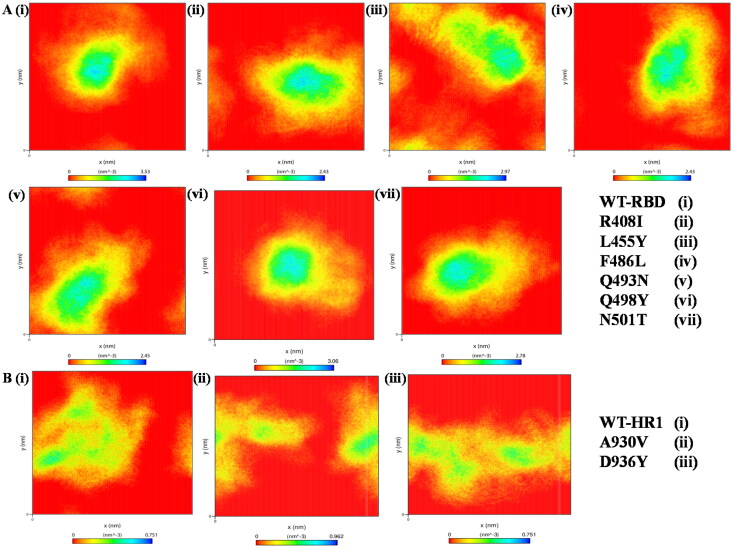
The simulation trajectories were further investigated to understand the atomic density distribution between the WT and mutations of both domains of spike glycoprotein to check the changes in atomic orientation and distribution on systems, plotted using densmap of GROMACS package (Hess et al., 2008). The results revealed that a stable partial density area was observed for WT within 3.5 nm−3 for RBD whereas decreased density was noticed for R408I, L455Y, F486L, Q493N, Q498Y, N501T ranging from 2.4 to 3.0 nm−3 (Figure 10A(i–vii)). The density area of each atom within the structure of mutant proteins of the HR1 domain was WT (0.7nm-3) and found to be 0.9-3 for A930V, 0.7nm-3 for D936Y (Figure 10B(i–iii)). The results indicate that the protein becomes less stable and displays multiple minimum energy wells in certain cases like L455Y in RBD and all the HR1 mutants.
Figure 10.
Density distribution of both RBD and HR1 domains with WT and mutant systems throughout MD simulations (A) RBD region of spike glycoprotein (i) WT, (ii-vii) Mutations of RBD region (B) HR1 domain (i) WT, (ii & iii) Mutations on mapped on HR1 domain.
Frustration analysis of WT vs mutants
The minimal frustrations of the spike glycoprotein associated RBD and HR1 domains are well characterized by the global energy minima occurrence with local minima. The results showed maximum frustrations leading to loss of contacts was noticed from ∼400-450 and ∼450-510 and around ∼600 residues, where RBD mutations were positioned. The gain of frustration was noticed on the RBD WT structure (Supplemental Figure S6A(i–vii). Consequently, HR1 mutants A930V and D936Y revealed high alterations between the residues ∼910-920 and ∼930-940 explored loss of contacts between HR1 domain residues on spike crude structure (Supplemental Figure S6B(i–iii)). From the frustration analysis, it was confirmed that the prioritized eight mutations (6-RBD and 2-HR1) showed an unsteady shift in the patterns at substitution positions of both RBD and HR1 domains were probably leading to un-stability and dysfunctional effect on SARS-CoV-2 spike glycoprotein.
Discussion
Our study initiated with computational predictions to understand the effect of mutations mapped on RBD and HR1 domains of SARS-CoV-2 spike protein. The in silico methods revealed that the selected mutations R408I, L455Y, F486L, Q493N, Q498Y, N501T on RBD, and A930V, D936Y on HR1 are highly deleterious, damaging and majorly responsible for the destabilization of spike glycoprotein.
MD simulations of the mutants also confirm the same as the RMS deviations, RMS fluctuations reveal that the presence of mutations on the RBD region and HR1 domain of SARS-CoV-2 result in loss of stability of the overall protein structures. A previous study has been reported that the RBD spike glycoprotein residues L455, F486, Q493, and N501 form major binding with the human ACE2 receptor (Wan et al., 2020). Interestingly, in our study too, the RBD mutations L455Y, F486L, Q493N, and N501T were observed to have the highest deviations as compared to the WT structure. These alterations in the mutant structures revealed the destabilizing effect on the RBD structure contributing to a flexible binding interface on the spike glycoprotein receptor binding site. The analysis of the intramolecular hydrogen bonding profile revealed the least number of the bonds in mutant structures F486L, and Q498Y, whereas the mutations R408I, L455Y, Q493N, and N501T projected average intra-molecular hydrogen bonds when compared to WT. The least number of hydrogen bonds and alterations in the bond formation of mutants reflect protein dysfunction which in turn disturbs the structure dynamics, stability, and compactness of WT spike glycoprotein. The MD simulations of HR1 domain mutations also reveal that the mutations are responsible for the destabilization of the WT structure of the spike glycoprotein. The analysis of Rg during MD simulations suggests an increase in overall dimensions and a decrease in surface area of spike glycoprotein with the presence of RBD and HR1 mutations which is leading to protein instability. Further, the alterations in conformational dynamics, frustration in residues, magnitude-motion of atoms and FEL analysis were observed by the presence of mapped mutations also demonstrated a significant effect on the spike glycoprotein de-stabilization in comparison to the WT.
The mutant R408I on the RBD domain persists a positive charge (Arg) which is substituted by an aliphatic uncharged (Iso) residue, this change in charge can affect the receptor binding. Most of the mutations like F486L, Q493N, L455Y, and Q498Y are known to interact and N501T was found in forming a hydrogen bond with ACE2 in the viral entry to the host cells. The binding between the surface of RBD and ACE2 is having a coiled conformation, which is susceptible to changes by the mutations too. Our MD simulations also confirm the de-stabilizing effects of the mutations which change its amino acid properties and alter the binding affinity against ACE2 of human, which can potentially affect the efficiency of viral entry to host cells (Brielle et al., 2020; Lokman et al., 2020; Ortega et al., 2020; Ou et al., 2020; Perilla et al., 2015; Shang, Wan, et al., 2020). The A930V and D936Y mutations of the HR1 domain play a significant role in the infection process as the domain is vital for the viral membrane fusion and host cell entry forming the helical bundles. Existing reports stated that the substitution of Ala to Val is one of the major causes of resistance in the mouse hepatitis coronavirus which was studied against HR1 and HR2 derived peptide inhibitors (Bosch et al., 2008), the spike protein mutation A930V of HR1 may also have a similar effect, which can be investigated further. The outcomes of the current study potentially have implications for research focused on targeting the spike protein- ACE2 receptor binding site for the development of novel therapeutics and vaccines to mitigate COVID-19 infections. The MD simulations also enhance our molecular-level understanding of the dynamic effects of the mutations on the prefusion spike glycoprotein RBD and HR1 domain, which affects the binding.
Contribution of authors
SA contributed to literature mining, extensive in silico exercises, MD simulations investigations, and write-up in the manuscript. HK contributed to write-up and corrections in the manuscript. DG contributed to the designing the hypothesis, major inputs, correspondence, and entire corrections in the manuscript.
Supplementary Material
Funding Statement
SA is thankful for the Research Associate Fellowship by the Indian Council of Medical Research (ICMR), India, Project ID: (2019-6039 File No. ISRM/11(83)/2019). HK is the recipient of DBT-BioCARe women scientist fellowship awarded by the Department of Biotechnology, India (No.BT/PR31715/BIC/101/1233/2019). DG acknowledges the funding by the Department of Biotechnology, Government of India (Grant No. BT/BI/25/001/2006).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Adzhubei, I., Jordan, D. M., & Sunyaev, S. R. (2013). Predicting functional effect of human missense mutations using PolyPhen-2. Current Protocols in Human Genetics , 76(1), 7.20.1–7.20.41. 10.1002/0471142905.hg0720s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahamad, S., Hassan, M. I., & Dwivedi, N. (2018). Designing of phenol-based β-carbonic anhydrase1 inhibitors through QSAR, molecular docking, and MD simulation approach. 3 Biotech, 8(5), 256. 10.1007/s13205-018-1278-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahamad, S., Islam, A., Ahmad, F., Dwivedi, N., & Hassan, M. I. (2019). 2/3D-QSAR, molecular docking and MD simulation studies of FtsZ protein targeting benzimidazoles derivatives. Computational Biology and Chemistry, 78, 398–413. 10.1016/j.compbiolchem.2018.12.017 [DOI] [PubMed] [Google Scholar]
- Amir, M., Ahamad, S., Mohammad, T., Jairajpuri, D. S., Hasan, G. M., Dohare, R., Islam, A., Ahmad, F., & Hassan, M. I. (2019). Investigation of conformational dynamics of Tyr89Cys mutation in protection of telomeres 1 gene associated with familial melanoma. Journal of Biomolecular Structure and Dynamics, 26, 1–15. [DOI] [PubMed] [Google Scholar]
- Amir, M., Ahmad, S., Ahamad, S., Kumar, V., Mohammad, T., Dohare, R., Alajmi, M. F., Rehman, T., Hussain, A., Islam, A., Ahmad, F., & Hassan, M. I. (2020). Impact of Gln94Glu mutation on the structure and function of protection of telomere 1, a cause of cutaneous familial melanoma. Journal of Biomolecular Structure & Dynamics, 38(5), 1514–1524. 10.1080/07391102.2019.1610500 [DOI] [PubMed] [Google Scholar]
- Andersen, K. G., Rambaut, A., Lipkin, W. I., Holmes, E. C., & Garry, R. F. (2020). The proximal origin of SARS-CoV-2. Nature Medicine, 26(4), 450–452. 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch, B. J., Rossen, J. W., Bartelink, W., Zuurveen, S. J., de Haan, C. A., Duquerroy, S., Boucher, C. A., & Rottier, P. J. (2008). Coronavirus escape from heptad repeat 2 (HR2)-derived peptide entry inhibition as a result of mutations in the HR1 domain of the spike fusion protein. Journal of Virology, 82(5), 2580–2585. 10.1128/JVI.02287-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brielle, E. S., Schneidman-Duhovny, D., & Linial, M. (2020). The SARS-CoV-2 exerts a distinctive strategy for interacting with the ACE2 human receptor. Viruses, 12(5), 497–417. 10.3390/v12050497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella, M., Rajnik, M., Cuomo, A., Dulebohn, S. C., & Di Napoli, R. (2020). Features, evaluation and treatment coronavirus (COVID-19). In Babak Abai et al. (Eds.), StatPearls. StatPearls Publishing. 10.2214/AJR.20.22954 [DOI] [PubMed] [Google Scholar]
- Choudhury, A., & Mukherjee, S. (2020). In silico studies on the comparative characterization of the interactions of SARS‐CoV‐2 spike glycoprotein with ACE‐2 receptor homologs and human TLRs. Journal of Medical Virology, 92, 1–9. 10.1002/jmv.25987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard, B., Valle, C., de Lamballerie, X., Canard, B., Seidah, N. G., & Decroly, E. (2020). The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Research, 176, 104742. 10.1016/j.antiviral.2020.104742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappier, V., Chartier, M., & Najmanovich, R. J. (2015). ENCoM server: Exploring protein conformational space and the effect of mutations on protein function and stability. Nucleic Acids Research, 43(W1), W395–W400. 10.1093/nar/gkv343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giurgea, L. T., Han, A., & Memoli, M. J. (2020). Universal coronavirus vaccines: The time to start is now. NPJ Vaccines, 5(1), 1–3. 10.1038/s41541-020-0198-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex, N., & Peitsch, M. C. (1997). SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis, 18(15), 2714–2723. 10.1002/elps.1150181505 [DOI] [PubMed] [Google Scholar]
- Hess, B., Kutzner, C., Van Der Spoel, D., & Lindahl, E. (2008). GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of Chemical Theory and Computation, 4(3), 435–447. 10.1021/ct700301q [DOI] [PubMed] [Google Scholar]
- Jia, Y., Shen, G., Zhang, Y., Huang, K. S., Ho, H. Y., Hor, W. S., Yang, C.-H., Li, C., & Wang, W. L. (2020). Analysis of the mutation dynamics of SARS-CoV-2 reveals the spread history and emergence of RBD mutant with lower ACE2 binding affinity. bioRxiv. https://doi.org/ 10.1101/2020.04.09.034942 [DOI] [Google Scholar]
- Johansson, M. U., Zoete, V., Michielin, O., & Guex, N. (2012). Defining and searching for structural motifs using DeepView/Swiss-PdbViewer. BMC Bioinformatics, 13, 173. 10.1186/1471-2105-13-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanipakam, H., Sharma, K., Thinlas, T., Mohammad, G., & Pasha, M. Q. (2020). Structural and functional alterations of Nitric Oxide Synthase 3 due to missense variants associate with high-altitude pulmonary edema through dynamic study. Journal of Biomolecular Structure and Dynamics, 14, 1–16. [DOI] [PubMed] [Google Scholar]
- Lan, J., Ge, J., Yu, J., Shan, S., Zhou, H., Fan, S., Zhang, Q., Shi, X., Wang, Q., Zhang, L., & Wang, X. (2020). Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature, 581(7807), 215–216. 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- Lokman, S. M., Rasheduzzaman, M., Salauddin, A., Barua, R., Tanzina, A. Y., Rumi, M. H., Hossain, M. I., Siddiki, A. Z., Mannan, A., & Hasan, M. M. (2020). Exploring the genomic and proteomic variations of SARS-CoV-2 spike glycoprotein: A computational biology approach. bioRxiv. https://doi.org/ 10.1101/2020.04.07.030924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega, J. T., Serrano, M. L., Pujol, F. H., & Rangel, H. R. (2020). Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: An in silico analysis. EXCLI Journal, 19, 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, J., Zhou, Z., Zhang, J., Lan, W., Zhao, S., Wu, J., Seto, D., Zhang, G., & Zhang, Q. (2020). RBD mutations from circulating SARS-CoV-2 strains enhance the structural stability and human ACE2 affinity of the spike protein. bioRxiv. https://doi.org/ 10.1101/2020.03.15.991844 [DOI] [Google Scholar]
- Pandurangan, A. P., Ochoa-Montaño, B., Ascher, D. B., & Blundell, T. L. (2017). SDM: A server for predicting effects of mutations on protein stability. Nucleic Acids Research, 45(W1), W229–W235. 10.1093/nar/gkx439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra, R. G., Schafer, N. P., Radusky, L. G., Tsai, M. Y., Guzovsky, A. B., Wolynes, P. G., & Ferreiro, D. U. (2016). Protein Frustratometer 2: A tool to localize energetic frustration in protein molecules, now with electrostatics. Nucleic Acids Research, 44(W1), W356–W360. 10.1093/nar/gkw304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilla, J. R., Goh, B. C., Cassidy, C. K., Liu, B., Bernardi, R. C., Rudack, T., Yu, H., Wu, Z., & Schulten, K. (2015). Molecular dynamics simulations of large macromolecular complexes. Current Opinion in Structural Biology, 31, 64–74. 10.1016/j.sbi.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires, D. E., Ascher, D. B., & Blundell, T. L. (2014). DUET: A server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Research, 42(Web Server issue), W314–W319. 10.1093/nar/gku411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires, A. S., Porto, W. F., Franco, O. L., & Alencar, S. A. (2017). In silico analyses of deleterious missense SNPs of human apolipoprotein E3. Scientific Reports, 7(1), 1–9. 10.1038/s41598-017-01737-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk, S., Páll, S., Schulz, R., Larsson, P., Bjelkmar, P., Apostolov, R., Shirts, M. R., Smith, J. C., Kasson, P. M., van der Spoel, D., Hess, B., & Lindahl, E. (2013). GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics (Oxford, England)), 29(7), 845–854. 10.1093/bioinformatics/btt055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues, C. H., Pires, D. E., & Ascher, D. B. (2018). DynaMut: Predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Research, 46(W1), W350–W355. 10.1093/nar/gky300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang, P., Du, X., Yang, L.-Q., Meng, Z.-H., & Liu, S.-Q. (2017). Molecular motions and free-energy landscape of serine proteinase K in relation to its cold-adaptation: A comparative molecular dynamics simulation study and the underlying mechanisms. RSC Advances, 7(46), 28580–28590. 10.1039/C6RA23230B [DOI] [Google Scholar]
- Schreiner, E., Trabuco, L. G., Freddolino, P. L., & Schulten, K. (2011). Stereochemical errors and their implications for molecular dynamics simulations. BMC Bioinformatics, 12, 190. 10.1186/1471-2105-12-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede, T., Kopp, J., Guex, N., & Peitsch, M. C. (2003). SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Research, 31(13), 3381–3385. 10.1093/nar/gkg520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, J., Wan, Y., Luo, C., Ye, G., Geng, Q., Auerbach, A., & Li, F. (2020). Cell entry mechanisms of SARS-CoV-2. Proceedings of the National Academy of Sciences of the United States of America, 117(21), 11727–11734. 10.1073/pnas.2003138117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, J., Ye, G., Shi, K., Wan, Y., Luo, C., Aihara, H., Geng, Q., Auerbach, A., & Li, F. (2020. a). Structural basis of receptor recognition by SARS-CoV-2. Nature, 581(7807), 221–224. 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, J., Ye, G., Shi, K., Wan, Y., Luo, C., Aihara, H., Geng, Q., Auerbach, A., & Li, F. (2020. b). Structural basis for receptor recognition by the novel coronavirus from Wuhan. Research Square. https://doi.org/1021203/rs224749/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim, N. L., Kumar, P., Hu, J., Henikoff, S., Schneider, G., & Ng, P. C. (2012). SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Research, 40(Web Server issue), W452–W457. 10.1093/nar/gks539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X., Wu, C., Li, X., Song, Y., Yao, X., Wu, X., Duan, Y., Zhang, H., Wang, Y., Qian, Z., Cui, J., & Lu, J. (2020). On the origin and continuing evolution of SARS-CoV-2. National Science Review, 7(6), 1012–1023. 10.1093/nsr/nwaa036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, Y., Shang, J., Graham, R., Baric, R. S., & Li, F. (2020). Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. Journal of Virology, 94(7), e0012720. 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worth, C. L., Preissner, R., & Blundell, T. L. (2011). SDM-a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Research, 39(Web Server issue), W215–W222. 10.1093/nar/gkr363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., Graham, B. S., & McLellan, J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.).), 367(6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielkiewicz, J. (2005). Structural properties of water: Comparison of the SPC, SPCE, TIP4P, and TIP5P models of water. The Journal of Chemical Physics, 123(10), 104501. 10.1063/1.2018637 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.