Summary
Exophiala dermatitidis is a polyextremotolerant fungus with a small genome, thus suitable as a model system for melanogenesis and carotenogensis. A genome-scale model, iEde2091, is reconstructed to increase metabolic understanding and used in a shadow price analysis of pigments, as detailed here. Important to this reconstruction is OptFill, a recently developed alternative gap-filling method useful in the holistic and conservative reconstruction of genome-scale models of metabolism, particularly for understudied organisms like E. dermatitidis where gaps in metabolic knowledge are abundant.
For complete details on the use and execution of this protocol, please refer to Schroeder and Saha (2020) and Schroeder et al. (2020).
Graphical Abstract

Highlights
-
•
Detailed instructions are given for genome-scale reconstruction of E. dermatitidis
-
•
This protocol accomplishes reconstruction by making use of the OptFill tool
-
•
Detailed instructions are given to reproduce a shadow price analysis
-
•
Generic instructions are given to replicate this work for other organisms
Exophiala dermatitidis is a polyextremotolerant fungus with a small genome, thus suitable as a model system for melanogenesis and carotenogensis. A genome-scale model, iEde2091, is reconstructed to increase metabolic understanding and used in a shadow price analysis of pigments, as detailed here. Important to this reconstruction is OptFill, a recently developed alternative gap-filling method useful in the holistic and conservative reconstruction of genome-scale models of metabolism, particularly for understudied organisms like E. dermatitidis where gaps in metabolic knowledge are abundant.
Before You Begin
Timing: hours to weeks
This section includes procedure which is necessary to the successful completion of the steps detailed later in this protocol, including needed programming languages, suggested text editors and FTP protocols (if utilizing a computing cluster), and locations of repositories which contain code and files used throughout this work.
-
1.
If needed, install the latest version of programming languages used throughout this protocol. These languages are Perl, Python, and the General Algebraic Modeling System (GAMS).
-
a.
The Perl programming language can be downloaded from www.perl.org. Perl is standard on Mac OS and Linux/UNIX operating systems, but the authors still encouraged users to ensure that the latest version of Perl is installed to ensure compatibility with modules downloaded from the Comprehensive Perl Archive Network (CPAN) which are used throughout this protocol. For Windows operating system users, the Strawberry Perl download, which can be found at http://strawberryperl.com/, is recommended as this is what is used by the authors. All Perl language downloads are free of charge.
-
b.
The Python programming language can be downloaded from www.python.org. Similar to Perl, there are separate download packages for Windows, Mac OS, and Linux/UNIX. So be sure that the correct package is downloaded. All Python language downloads are free of charge.
-
c.
The programming language and platform used by the authors for optimization applications, such as OptFill, Flux Balance Analysis (FBA), and Flux Variability Analysis (FVA), is the GAMS which can be downloaded from www.gams.com. GAMS is not free of charge, as the base module and CPLEX solver must be purchased.
-
2.
A text editing software with capabilities beyond those of standard text editing software (e.g., Notepad for Windows or TextEdit for Mac OS) such as highlighting for programming languages is recommended. The authors, using Windows, use Notepad++, available from notepad-plus-plus.org. Those who use Mac OS in the authors’ laboratory use BBEdit, available from https://www.barebones.com/products/bbedit/, or Sublime Text, available from https://www.sublimetext.com/, for their text editing.
-
3.
In this protocol, the authors make use of a supercomputing cluster (the Holland Computing Center, HCC) to perform much of this protocol, including all code in the GAMS programming language. If the readers of this protocol have access to resources which will increase solution speed, such as a supercomputing cluster at their own institutions, follow that institution’s or supercomputing cluster’s recommendations on creating an account, accessing the cluster and filesystems, and any other pertinent recommendations. For managing files on the supercomputing cluster, the authors have used the nppFTP (Notepad plus plus File Transfer Protocol) plugin as the authors use the Windows operating system. For connecting to the supercomputing cluster, the authors use PuTTY (compatible with Windows and Unix) which can be found at https://www.chiark.greenend.org.uk/∼sgtatham/putty/. For FTP needs, Mac OS users in the authors’ laboratory use Cyberduck, available at https://cyberduck.io/.
-
4.
Codes related to this protocol as well as to Schroeder and Saha (2020) and Schroeder et al. (2020) can be found in two GitHub repositories associated with these works: GitHub repositories for both the iEde2091 model (doi:10.5281/zenodo.3608172, also available and periodically updated on GitHub at https://github.com/ssbio/E_dermatitidis_model) and OptFill (doi: 10.5281/zenodo.3518501, also available and periodically updated on GitHub at https://github.com/ssbio/OptFill).
-
5.
For many of these protocol steps, an internet connection is required.
-
6.
It should be noted that here screenshots with some shapes overlaid are used to help guide users through the steps of this protocol; however, these screenshots were taken in April of 2020, whereas the actual process of building the iEde2091 model began as early as March of 2017. Therefore, this protocol is not an exact replication of what was done to construct the iEde2091 model, but rather demonstrates the steps of model reconstruction.
Collecting and Compartmentalizing Known Metabolic Functions of the Understudied Organism, Exophiala dermatitidis
Timing: days to months
As discussed in Schroeder et al. (2020), Exophiala dermatitidis is a highly melanized polyextremotolerant fungi which is cultural as both a yeast and as a mycellium. Due to this and its small genome, it is of interest as a model polyextremotolerant organism and model defensive pigment producing organism; however, lack of genome annotation results in poor knowledge related to metabolic functions and their compartmentalization. This is addressed through utilizing multiple data sources, predictive tools, and knowledge of related organisms. This portion of the protocol is focused on the creation of the first draft model of the iEde2091 model, as an essential precursor for having a sufficient metabolic model draft to which to apply OptFill. This method is used in Schroeder et al. (2020) for model reconstruction.
-
7.
Get information as to proteins and genes known to be in Exophiala dermatitidis (UniProt). Perform a basic search of the UniProt database (uniprot.org) using the phrase “Exophiala dermatitidis” (gold oval in Figure 1). The results should look like what is shown in Figure 1. Important features of the resultant screen are highlighted with colored ovals or arrows.
-
a.
Select the “columns” button (pink oval) which allows addition or removal of certain columns in the output table.
-
b.
Once selected, a large menu will appear with the header “Customize results table”. In the menu, one drop-down menu should have the header “Function” (see Figure 1). Select the Enzyme Commission (EC) number checkbox under that header, and then select the button “Save” near the bottom of the webpage. In total, this should add one column with the header “EC number” at the position of the pink arrow in Figure 1.
-
c.
Next, select the “Download” button (orange oval in Figure 1). This will open a menu. From the “Format” drop-down menu select “Excel”, select the radio button next to “Uncompressed”, and finally select “Go”. This will result in a downloaded Microsoft Excel table.
-
8.
Remove false search results (UniProt). It is possible that some of the results downloaded from the previous step include proteins not from Exophiala deramtitidis. These entries will be removed in this step. They can be most easily removed by sorting the Excel file from step 7 by the “organism” column a deleting all rows which do not list Exophiala dermatitidis.
-
9.
Expand the known number of EC Numbers (UniProt). The EC number provides a link between the gene and/or protein and the metabolic function(s) which the resultant enzyme carries out. However, this crucial link is missing for many of these protein entries. This step will use Perl language code developed by the authors and input files developed in step 7, which can be found in the GitHub repository associated with Schroeder et al. (2020) (doi: 10.5281/zenodo.3608172). Both code and inputs to the code are indicated by the dark red arrows as shown in Figure 2, namely “UniProt_get_ECs.pl” and “Uniprot_E_dermatitidis.csv”. This code utilizes the BRENDA database to fill in gaps in the knowledge of EC number and protein links, and requires the file indicated by the pink arrows on Figure 2. Note that for most code in the GitHub repository associated with Schroeder et al. (2020), the “common_functions.pl” file is necessary as it is a library of Perl language functions utilized by many codes (indicated by pink arrow).
Figure 1.
Screenshot of UniProt Search for Exophiala dermatitidis Proteins with Interesting Features Highlighted
The gold oval highlights the search query used, the pink ovals highlights where options may be found to expand the table to include Enzyme Commission (EC) numbers as an additional column, and the orange oval indicates where the table of results may be downloaded in the form of an Excel table. The excerpt on the right is a partial screenshot of the menu which appears when the button in either pink oval is selected. The pink arrow indicates the checkbox which should be selected.
Figure 2.
Screenshot of the Files Available
The screenshot is from the database associated with Schroeder et al. (2020) as of July 15, 2020. Arrows are used to highlight codes referred to in various steps of this protocol, with the color of those arrows indicating the groupings of the codes. Dark red and pink arrows are files associated with Before You Begin step 9; orange and pink arrows with Before You Begin step 10; yellow arrows with Before You Begin steps 13 and 15, as well as Step-by-Step Methods steps 1; brown arrows with Before You Begin step 13; green arrows with Step-by-Step Methods step 27; light blue arrows with Before You Begin step 14 and Step-by-Step Methods step 13; dark blue arrows with Step-by-Step Methods step 19; and purple arrows with Step-by-Step Methods step 31. While not a significant part of this protocol, python-based code has been provided so that users who do not have access to GAMS can perform some basic analyses on the iEde2091 model including FBA, FVA, and shadow price analyses (gray arrows). Note that the code “TXTtoSBMLmodel.pl” is also marked with a gray arrow as it converts iEde2091 to SBML format (resulting in “iEde2091.xml”).
-
a.
The code as provided requires a Comma-Separated Values (CSV) file as an input containing the information from UniProt. The table downloaded in step 7 should be saved as a “.csv” extension through Microsoft Excel by selecting “save as” and then selected “CSV (comma delimited) (∗.csv)” from the drop-down file type menu.
-
b.
Download, from the GitHub repository associated with Schroeder et al. (2020) the following codes: “UniProt_get_ECs.pl” (dark red arrow in Figure 2), “Uniprot_E_dermatitidis.csv” (dark red arrow in Figure 2, or use the file from step 7 and rename it as this), and “common_functions.pl” (pink arrow in Figure 2). All these files should be in the same pathway.
-
c.
For Windows users, open the command prompt, for Mac and Unix users, open the terminal. Navigate the working directory to where the files are located from step 7c. To run the code, simply type “perl Uniprot_get_ECs.pl >output.txt”. The “>output.txt” portion should place what would normally be written to the command prompt or terminal to a text file named “output.txt” for later viewing.
-
d.
The result should be two files: “output.txt” and “UniProtECno.csv”. The former should have line formatting along the lines of “Searching Brenda for |Protein Name|..EC:<list of EC numbers found>”. The latter should have the same formatting as “Uniprot_E_dermatitidis.csv”, yet with the results filled into the “EC number” column.
CRITICAL: The file “common_functions.pl” and the Perl module LWP (The World Wide-Web library for Perl available at https://metacpan.org/pod/LWP) are required for the automated BRENDA database search, otherwise the “Uniprot_get_ECs.pl” code will return an error before performing any function.
CRITICAL: A stable internet connection is required on the order of minutes to hours, depending on the size of the input excel sheet.
Note: Should the code as provided not work, consult the BRENDA webpage and compare it to the provided code, as BRENDA does not at present have a dedicated Application Programming Interface (API); therefore, the “UniProt_get_ECs.pl” works partially on the principle of “screen scraping” (e.g., reading the webpage, rather than the underlying database). Any code relying on this principle may be outdated, yet the current code should at least provide a base which can be updated.
Note: Generally, a warning will appear on the command line such as “Smartmatch is experimental at <pathway> line <number>”. This is normal and is to be expected. The experimental nature of Smartmatch has not, as of yet, caused any issues in code written by the authors which use this tool.
-
10.
Get information as to proteins and genes known to be in Exophiala dermatitidis (NCBI). The data used from the NCBI database can be accessed by the following series of substeps.
-
a.
Perform a search (orange arrow Figure 3A) from the NCBI homepage for “Exophiala dermatitidis”.
Figure 3.
Selection of Screenshots Intended to Guide Users to the Genome Assemble Data Used in This Protocol and to Download the Protein-Based Genome Annotation of the Selected Assembly
Screenshots taken on 04/20/2020.
(A) Search line query used.
(B) Screenshot of a portion of the results screen from that search line query showing where to find the genome assembly results.
(C) Partial screenshot of assembly results which highlights the assembly used in the creation of iEde2091 (Exop_derm_V1). This was the assembly chosen as it was the only assemble available as of 2017 when this part of the procedure was done for Schroeder et al. (2020). As of 04/20/2020 (when this screenshot was taken), there were seven genome assemblies.
(D) Partial screenshot of the genome assembly page, orange arrow shows the link to follow the assembly to the related organism page.
(E) Partial screenshot of the organism page, specifically the Entrez record table. Selecting the indicated link leads to a search results table.
(F) The search results table can be downloaded as a text-based table, which can be easily converted into a Microsoft Excel worksheet.
-
b.
Under the “Genomes” heading of the results, select the “Assembly” link (orange arrow Figure 3B).
-
c.
As this portion of the workflow was originally performed in 2017 for Schroeder et al. (2020), only one assembly was available at that time, which is the one labeled “Exop_derm_V1” (RefSeq assembly accession GCF_000230625.1, orange arrow Figure 3C) at present (as of March 2020). To recreate this work, this can be selected, or another assembly (such as the more recent ASM1088354v1 shown in Figure 3C) can be selected to improve upon this work or expand it.
-
d.
Follow the hyperlink after “Organism name: ” (generally the first hyperlink under the header, orange arrow in Figure 3D). This will take the reader to NCBI’s Taxonomy browser which was used for the creation of the iEde2091 model.
-
e.
In a table on the RHS of the web page, the number of direct links in the row “Gene” should be hyperlinked (orange arrow Figure 3E), following this hyperlink leads to a tabular list of genes in the genome assembly.
-
f.
This list can be downloaded as a table by selecting the “Send to:” drop-down menu above the upper right-hand corner of the results table.
-
i.
Select the radio button “file”, which will expand this menu.
-
ii.
Select “Tabular (text)”.
-
iii.
Sort by any method.
-
iv.
Select “Create File” (orange arrow Figure 3F).
Note: The formatting of the resultant file is moderately different from that of the supplemental files in Schroeder et al. (2020), as these files were originally generated in 2017.
-
11.
Expand the known number of EC Numbers (NCBI). This step strongly parallels step 9, and thus, for critical portions of this step and notes related to this step, see step 9. A similar set of input codes is used in this step, which can also be found in the GitHub repository associated with Schroeder et al. (2020), shown in Figure 2. The necessary codes for this step are indicated by orange arrows (“NCBIproteindetails.csv” and “NCBI_get_ECs.pl”) with the necessary library of functions indicated by the pink arrow (“common_functions.pl”).
-
a.
The first step for the recreation of the iEde2091 model is to convert the Microsoft Excel file to a .csv file using the procedure described in step 9a.
-
b.
Download, from the GitHub repository associated with Schroeder et al. (2020) the following codes: “NCBI_get_ECs.pl” (orange arrow in Figure 2), “NCBIproteindetails.csv” (orange arrow in Figure 2, or use the file from step 10 and rename it as this), and “common_functions.pl” (pink arrow in Figure 2, if not already downloaded). All these files should be in the same pathway (preferably the same as was used in step 9).
-
c.
For Windows users, open the command prompt, for Mac and Unix users, open the terminal. Navigate the working directory to where the files are located from step 9b. To run the code, simply type “perl NCBI_get_ECs.pl >output.txt”. The “>output.txt” portion should place what would normally be written to the command prompt or terminal to a text file named “output.txt” for later viewing.
Note: See critical notes and notes for step 9.
-
d.
The result should be two files: “output.txt” and “NCBIECno.csv”. The former should have line formatting along the lines of “Searching Brenda for |Protein Name|..EC:<list of EC numbers found>”. The latter should have the same formatting as “NCBIproteindetails.csv”, yet with the results filled into the “EC number” column.
-
12.
Determine a unique list of EC Numbers in Exophiala dermatitidis. The previous steps have developed a set of EC numbers linked to proteins and genes through the use of NCBI and UniProt databases. These databases have some overlap, and some uniqueness (for instance, the databases have a different number of proteins associated with Exophiala dermatitidis). These results then should be combined to get a full set of EC numbers known to be in E. dermatitidis. This is done by copying-and-pasting of the EC columns of the files generated in steps 9 and 11, and then performing the “remove duplicates” data operation on that column in Microsoft Excel. Now that metabolic functions known to be carried out by Exophiala dermatitidis have been identified, subcellular localization of those functions is necessary for modeling.
-
13.
Determining reactions associated with EC numbers through KEGG API. EC number can be used to link proteins to their corresponding metabolic functions. In this work, the KEGG Application Programming Interface (API, available at rest.kegg.jp) was used by the “Enzymes_to_rxns.pl” Perl language code (yellow arrow in Figure 2). Specifically, the “link” operation was used to link EC number to the reaction(s) which that EC number might catalyze through syntax such as “rest.kegg.jp/link/reaction/1.1.1.1” for EC number 1.1.1.1 (alcohol dehydrogenase). The “Enzymes_to_rxns.pl” code uses a text list of EC numbers, for example “EClist_1.txt” in Figure 2 (yellow arrow), and outputs a csv file where the first column is EC numbers and the second is KEGG reactions associated with those EC numbers.
Note: At this point, a non-KEGG system of identifiers could be chosen to use for this protocol, if so desired, which may be partially automated. One such example conversion is included in the GitHub for the iEde2091 model (https://github.com/ssbio/E_dermatitidis_model, and seen in Figure 2) which converts KEGG identifiers to BIGG identifiers at a rate of about 38% for reactions and 62% for metabolites, and the remainder would require manual curation to affect the conversion. The required files for this conversion are marked with brown arrows in Figure 2.
-
14.
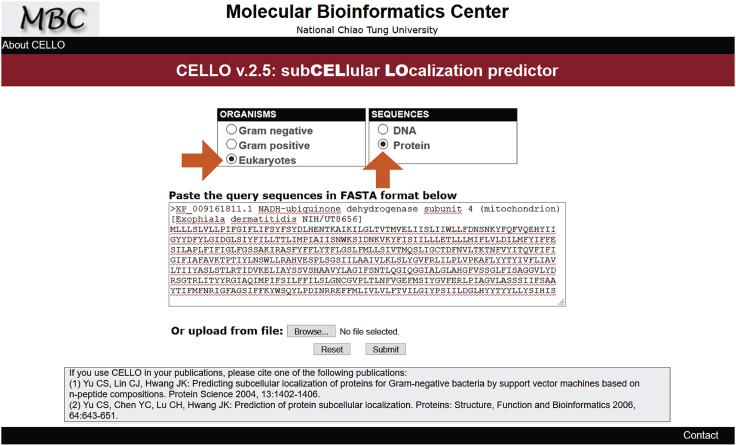
Determining compartmentalization of EC numbers through CELLO. Since Exophiala dermatitidis is an understudied organism, it was decided to use the CELLO v.2.5: subCELlular LOcalization predictor (available at cello.life.nctu.edu.tw) to determine compartmentalization of reactions through amino acid sequences of proteins associated with EC numbers (Yu et al., 2006). In these predictions, the “Eukaryotes” and “Proteins” radio button options were selected as shown in Figure 4 (orange arrows) which contains an example amino acid sequence for Exophiala dermatitidis. The amino acid sequences were retrieved as FASTAs from either UniProt or NCBI database, depending on the EC number for which compartmentalization is being investigated. This step was performed manually as CELLO lacks an Application Programming Interface (API) which would allow for automation. From this, a list of enzyme classification numbers with subcellular localizations appended (for example, “1.1.1.1[c]” for alcohol dehydrogenase in the cytosol) is generated as an input to the next step, named “ECs_with_comp.txt”.
Figure 4.
Screenshot of the CELLO Webpage and an Example Exophiala dermatitidis Enzyme Amino Acid Sequence Placed in the Query Box
Orange arrows indicate the settings to be selected. CELLO is used in Before You Begin step 14 to determine compartmentalization of the enzymes identified in Exophiala dermatitidis.
-
15.
Mapping EC numbers to a list of reactions. Next, the links between enzymes and reactions that EC numbers represent must be followed to get a list of reactions which can be catalyzed by enzymes known to be in Exophiala dermatitidis. The code “Enzymes_to_rxns.pl” (yellow arrow Figure 2) was used then to get list a list of reactions which might be catalyzed by these enzymes using “EC_with_comp.txt” as an input file. The resultant list of reaction identifiers and stoichiometries had reaction compartments applied automatically by the code used. This provides a list of reactions with stoichiometries in the same format used by the iEde2091’s model file.
-
16.
Selecting related species from which to fill metabolic gaps. An analysis of the reaction list generated by the previous step showed many metabolic gaps, even in key metabolic pathways such as the TCA cycle. Therefore, as done in other genome-scale modeling efforts, genome-scale models of closely related species were sought to fill these metabolic gaps. This search should be conducted using the most complete phylogenetic tree which could be found which includes the species of interest, at the time of this work the most complete phylogenetic tree for Ascomycota fungi is from Schoch et al. (2009), to identify closely related species. Google Scholar should be used to identify published genome-scale models of these species (if available). In Schroeder et al. (2020), the Aspergillus genus was identified as species most related to Exophiala dermatitisdis which had multiple published genome-scale models. Identified at the time were four metabolic models of Aspergillus species: A. nidulans by David et al. (2008), A. niger by Andersen, Nielsen and Nielsen (2008), A. oryzae by Vongsangnak et al. (2008), and A. terreus by Liu et al. (2013).
Note: Saccharomyces species, specifically the model species Saccharomyces cerevisiae (bakers’ yeast) were not selected as models from which to fill these metabolic gaps as ancestors of Saccharomyces and Exophiala species branched very early in the evolutionary history of Ascomycota fungi (Schoch et al., 2009). While a model of Saccharomyces cerevisiae was used in later manual curation efforts, namely iSce926 by Chowdhury, Chowdhury and Maranas (2015), Saccharomyces species were not considered sufficiently related for wide-scale adoption of metabolic functionalities.
-
17.
Download and compare selected Aspergillus models. The supplemental files associated with each publication of genome-scale model of a related species, in this work from the Aspergillus genus, identified in step 16 should then be downloaded. While each genome-scale modeling work had unique formatting, most models will list EC numbers associated with either the model or reaction which can then be used to compare the metabolic functionalities of each model. In Schroeder et al. (2020) all four models listed EC numbers associated with reactions. Often, through some relatively simple string handling in Microsoft Excel, lists of all EC numbers both with and without compartmentalization can be generated for each model, in addition to a non-compartmentalized list of all enzymes present in the four models in total. The non-compartmentalized enzyme list can be used to determine which enzymes are present in which models to generate a summary of the overlaps of the metabolic functionalities of the related genome-scale models used. In Schroeder et al. (2020), this overlap analysis resulted in four “bins” of EC numbers. The first bin was the full consensus (enzymes common to all four species models) which were assumed to also be present in E. dermatitidis. The remaining bins were labeled common to three of four, common to two of four, and unique to one model. These three bins will be used later to generate databases of functionalities for OptFill in steps 28, 32, and 36.
-
18.
Determine compartmentalization from Aspergillus models. For the full consensus bin from the previous step, each model should then be consulted to determine what compartmentalization these enzymes have in these models. All compartmentalization of the consensus enzymes found in the related speceis models was added to a list of enzymes with compartmentalization appended in the same manner as “EC_with_comp.txt” to produce a new list “Asp_EC_with_comp.txt”.
-
19.
Mapping EC numbers to a list of reactions (Aspergillus enzymes). The code “Enzymes_to_rxns.pl” (yellow arrow Figure 2) from step 15 should then be used to get a list of reactions which might be catalyzed by these enzymes using “Asp_EC_with_comp.txt” as an input file. The resultant list of reaction identifiers and stoichiometries should have reaction compartments applied automatically by the code. This step results in a list of reactions with stoichiometries in the same format used by model files such as iEde2091’s model file.
-
20.
Defining the biomass function. At this stage of the reconstruction process, literature evidence should be sought for the organism modeled in order to define a biomass equation, in this instance for Exophiala dermatitidis. Due to the availability of in vivo evidence for biomass composition at the time of developing the model, this search was divided primarily into three tasks: i) determine the composition of the cell wall and ii) determine the composition of the remainder of the cell (e.g., the plasma membrane and all it encloses) and iii) the mass fraction of biomass accounted for by carotenoids, where each accounts for a fraction of the total biomass. This method of defining the biomass composition was necessary due to direct evidence for the composition of E. dermatitidis cell walls being known, yet no evidence was yet present for the composition of the remainder of the cell or mass fraction of carotenoids in biomass. For the biomass of the cell wall, it was assumed that 25% of the cell mass is cell wall as the cell wall of this species has been described as “thick” (Chen et al., 2014; Kumar and Vatsyayan, 2010; Schnitzler et al., 1999) and in other ascomycetes cell walls may account for as much as 30% of dry biomass (Lipke and Ovalle, 1998). Schroeder et al. (2020) began with determining the composition of the cell wall. The composition of the cell wall of E. dermatitidis was obtained from Geis (1981). As A. terreus model is the most recently published of the four Aspergillus models, its biomass composition was used for the remainder cell (Liu et al., 2013). The lipid composition of the remainder of the cell was determined from A. terreus (by weight percentage) composition obtained from Kumar and Vatsyayan (2010). Once these two pieces of information were put together, one crucial piece of information was still missing: the contribution of carotenoids to the biomass of E. dermatitidis, as Aspergillus species lack carotenogensis. Again, lack of information pertaining to E. dermatitidis led to using the contribution of carotenoids to organism biomass data from another organism, in this case, Podospora anserina, for which approximately 3.47% of cell mass is carotenoids (Strobel et al., 2009). Both P. anserina and Exophiala are Ascomycota, but phylogenetically diverge at the class level. Unfortunately, no similar data was able to be found for a species which was phylogenetically closer to Exophiala (the search made was similar to that in step 16). The remaining cell weight, 71.53%, was assumed to be composed of the cell membrane and all biomass components enclosed within it (such as proteins and lipids). The stoichiometric ratio of these pseudometabolites in the biomass reaction was determined by using the solver tool in Microsoft Excel whose objective was a biomass molecular weight of 1,000 mg/gDW·h. Ratios between pseudometabolites were enforced in this analysis to preserve ratios as described above.
CRITICAL: The biomass pseudomolecule’s molecular weight should be 1,000 mg/gDW·h, so that the biomass flux rate simplifies to . Should this not be the case, the model will produce growth rates which are too fast or too slow, depending on the direction of error in the molecular weight of the biomass pseudomolecule. For a discussion on issues which can be caused by non-standard biomass weight, see Chan et al. (2017).
-
21.
Creating the first draft model. At this step, there are the results from three previous steps, namely steps 15, 19, and 20, which need to be synthesized for the creation of a genome-scale model. The results of step 15 contains reactions known to be able to be catalyzed in Exophiala dermatitidis; the results of step 19 contain a set of metabolic functions core to the related Aspergillus genus and assumed to also be core to E. dermatitidis; and step 20 produced the biomass pseudoreaction necessary for genome-scale models. Combining all three of these outputs, a first draft genome-scale model of Exophiala dermatitidis.
-
a.
This first draft E. dermatitidis model, or any other model made by paralleling this procedure, cannot be directly utilized by any code in the GAMS programming language, which requires precise formatting for input files. The set of files required by GAMS to properly read the model are generated by running the “convert∗.py” code (where “∗” is a short string identifier which describes what is being converted).
Note: The “convert∗.py” code generates several files which are GAMS-readable inputs including “mets.txt” (set of metabolites), “rxns.txt” (set of reactions), “rxntype.txt” (stores parameter specifying reaction direction), “Sij.txt” (stoichiometric matrix), “ex_rxns.txt” (subset of reactions that are exchange reactions), “irrev_rxns_f.txt” (subset of reactions which are irreversible and forward), “rev_rxns_no_ex.txt” (subset of reactions which are reversible not including exchange reactions), “irrev_rxns_b.txt” (subset of reactions which are irreversible backwards), and “reg_rxns.txt” (subset of reactions which are turned off by regulation). Only two copies of the “convert∗.py” code have been provided in the GitHub repository associated with Schroeder et al. (2020), as only line 20 need be edited to apply to a different model, and lines 23 through 32 can be edited to ensure that the output files of multiple “convert∗.py” codes do not write over eachother. This code can be run through Unix or Mac OS terminal or Windows Command Prompt using the command “python convert∗.py” so long as the “convert∗.py” code is contained in the current working directory.
-
b.
Once the “convert∗.py” code has been run on the correct model file, the FBA code “FBA.gms” can be run using the command “gams FBA.gms”. Users can change which model FBA is applied to by changing line 20 in the “convert∗.py” code and rerunning that code.
-
c.
At this step, this draft model will not be able to produce biomass, as there is no transport or exchange reactions. Further, there may be some Thermodynamically Infeasible Cycles (TICs), also called futile or type III cycles, due to lack of curation at this present stage, since all reactions at this point are reversible.
Manual Curation of the First Draft Exophiala dermatitidis Model
Timing: weeks to months
While the first draft of the Exophiala dermatitidis model has been created at this point, this first draft lacks important metabolic modeling capabilities such as the ability to produce the defensive pigments which make E. dermatitidis of such interest and the ability to simulate biomass production. Therefore, this portion of the protocol is focused on the creation of the second draft model of the iEde2091 model, which is capable of melanogenesis, carotenogenesis, and biomass production. This was done before the application of OptFill to ensure correct sysnthesis pathways for both melanins and carotenoids (rather than pathways predicted through OptFill). This second draft model is distinct from the first by having melanogenesis and carotenogensis added to the model and ensuring biomass can be produced. This procedure is used in Schroeder et al. (2020) in model reconstruction.
-
22.
Addition of melanogenesis. Since one of the goals of this study was the study of melanin synthesis (melanogenesis), the reaction pathways which produce melanins, namely pyomelanin, DHN-melanin, and eumelanin should be identified and added to the draft model at this stage. All reactions should be element and charge balanced as they are added to the model. In Schroeder et al. (2020) a primary source used for identify these reactions and genes related to melanin synthesis was Chen et al. (2014). From other available literature sources the details of melanin synthesis pathways, including DHN-melanin synthesis pathway (Szaniszlo (2002)), several types of fungal melanin (Eisenman and Casadevall (2012)), and detail pyomelanin synthesis in the related Apsergillus genus (Schmaler-Ripcke et al. (2009)) reactions and their associated stoichiometeries for the production of melanins were determined. An effort was made to identify as many compounds and reactions as possible with KEGG identifiers, however not all reactions and metabolites could be thus identified. For example, in the DHN-melanin synthesis pathway (showing melanins synthesized from dihydronaphthalene, abbreviated DHN) the final metabolite in the synthesis 1,8-DHN has no KEGG identifier, and thus was labeled “X00001”. As the metabolite identifier does not exist, neither do the reactions which produce 1,8-DHN and consume 1,8-DHN, therefore these reactions were arbitrarily assigned labels “R900” and “R901”. Compartmentalization for these reactions was obtained from Eisenman and Casadevall (2012).
-
23.
Addition of carotenogensis. Another particular goal of this study was to study the synthesis of carotenoids, therefore also the reaction pathways which produce melanins should be identified and added to the draft model at this stage. As in step 22, all reactions should be element and charge balanced as they are added to the model. In (Schroeder et al., 2020), carotenoids identified as producible by Exophiala dermatitidis were obtained from Geis and Szaniszlo (1984) and Kumar (2018), consisting primarily of -carotene, neurosporaxanthin, and β-apo-4′-carotenal (several other intermediate carotenoids are produced, but do not accumulate significantly). The carotenoid synthesis as described in Kumar (2018) was used to define the carotenoid synthesis pathway in the Exophiala dermatitidis model. Reaction stoichiometry, reaction labels, metabolite labels, and the synthesis pathway of the carotenoid precursor geranyl-geranyl-diphosphate were obtained from the KEGG database. These reactions were assumed to occur in the cytosol in the absence of concrete evidence as to compartmentalization.
-
24.
Addition of exchange reactions. At this point, the draft model is a collection of reactions, lacking any interactions with the environment outside the cell, and lacking interactions between the subcellular compartments. The first step in remedying this is the addition of exchange reaction which identifies how Exophiala dermatitidis interacts with its environment, specifically in terms of metabolites exchanged between the defined metabolic system (that is the cell, cell wall and nearby extracellular space) and the medium at large. As an aerobic organism, uptake of oxygen and export of carbon dioxide should be added. Further, it has been noted in Kumar (2018) through the defined medium used that glucose and sucrose may be used as a carbon sources, as well as sulfate may be used as a sulfate source, ammonium as a nitrogen source, and phosphate as a phosphorous source. Therefore, exchange reactions for these metabolites should also be added to the model. From discussions with an expert on Exophiala dermatitidis (namely our co-author Dr. Steven D. Harris), it was indicated that E. dermatitidis is also able to grow on acetate and ethanol as carbon sources as well, and that this should be reflected in the model, necessitating two more exchange reactions. Further, water and proton exchanges should be specified with the external environment.
Note: Not all exchange reactions present in the final model were defined here. For instance, it was discovered that some compound was necessary which allowed for the export of waste nitrogen. Here, nitrogen export is modeled as an export of uric acid.
-
25.
Addition of transport reactions. Now that step 24 has established interactions between Exophiala dermatitidis and its environment, interactions between the subcellular compartments (for instance, between the extracellular space and the cytosol) need to be defined using transport reactions. At this stage, some basic transport reactions should be added to the model which are very likely to exist in Exophiala dermatitidis, such as the diffusion of water across plasma membranes, the transport of exchanged metabolites from the extracellular space into the cytosol and to subcellular compartments which utilizes those resources (for instance, oxygen is transported into the mitochondria). Other exchange reactions which are common to the four chosen related species models from step 17 should also be considered for addition to the model at this stage depending on the presence of those metabolites in the current E. dermatitidis draft model.
Note: Not all transport reactions present in the final model were defined here, a significant number of transport reactions are added to the model as it is manually curated.
-
26.
Selection of metabolic functions used in manual curation. Even with the addition of exchange and transport reactions, the current Exophiala dermatitidis draft model has relatively few reactions which are capable of holding flux as determined by FVA, see “General steps on how to use iEde2091” and accompanying code for a description on how to apply FVA). This is due to the continued presence of a large number of metabolic gaps. Here, a database, or databases, of functionalities which can be used to manually fill metabolic gaps should be identified. In Schroeder et al. (2020), the databases selected were the iSce926 model (Chowdhury et al., 2015) and the set of enzymes common to three of four Aspergillus models from step 17. The latter database was converted to a set of reactions using the code “EC_to_rxns.pl”.
-
27.
Manual curation to ensure the production of biomass. The set of reactions selected in step 26 can then be used to curate the model were then used to manually used to curate the model such that the model can produce biomass. Manual curation can involve changing the direction of reactions already in the model, adding reactions from the database to address metabolic gaps noticed in the model, and removing reactions which may participate in TICs. In Schroeder et al. (2020) notes about what was done during manual curation can be found in “curation_notes.txt” in the GitHub repository associated with Schroeder et al. (2020) (green arrow in Figure 2). This curated model is the second draft model of Exophiala dermatitidis in this reconstruction. As noted in Schroeder et al. (2020), the model contains 1,587 reactions, of which only 711 are capable of holding flux.
Note: As defensive pigments are part of the biomass equation, this step also ensures that the defensive pigments which are being studied will also be produced.
Note: At this stage, the OptFill method could be used to address the metabolic gaps in the model using the iSce926 model and the set of reactions common to three of four Aspergillus models to define the database. This was not done at this stage in the original work because, at this point in time, the OptFill method had not yet been devised. After attempting to use GapFind/GapFill at this stage and finding that many metabolic gaps required multiple reactions to fix, it was desired by the authors to devise a method which could guarantee a minimized number of reactions added on a whole-model basis when gap-filling to achieve a conservative reconstruction (e.g., minimizing the global number of reactions added to the model to address a large number of metabolic gaps). This step in the reconstruction of an Exophiala dermatitidis model provided the rational and motivation for developing the OptFill method. As OptFill took more than a year to formulate, it was never applied at this stage since this formulation effort was done in parallel with the development of this genome-scale model of Eoxphiala dermatitidis. Details on the development of OptFill can be found in Schroeder and Saha (2020) and Schroeder et al. (2020).
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| National Center for Biotechnology Information (NCBI) | National Center for Biotechnology Information www.ncbi.nlm.nih.gov | RRID:SCR_006472 |
| Universal Protein Resource (UniProt) | UniProt Knowledgebase www.uniprot.org | RRID:SCR_002380 |
| Kyoto Encyclopedia of Genes and Genomes (KEGG) | Kyoto Encyclopedia of Genes and Genomes | RRID:SCR_012773 |
| GitHub | www.github.com | RRID:SCR_002630 |
| GitHub repository related to iEde2091 model | https://github.com/ssbio/E_dermatitidis_model | N/A |
| GitHub repository related to the OptFill tool | https://github.com/ssbio/OptFill | N/A |
| Experimental Models: Organisms/Strains | ||
| Aspergillus niger genome-scale model | Andersen, Nielsen and Nielsen (2008) | N/A |
| Aspergillus nidulans genome-scale model | David et al. (2008) | N/A |
| Aspergillus terreus genome-scale model | Liu et al. (2013) | N/A |
| Aspergillus oryzae genome-scale model | Vongsangnak et al. (2008) | N/A |
| Software and Algorithms | ||
| Protein Basic Local Alignment Search Tool (BLAST) | BLASTp https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins |
RRID:SCR_001010 |
| Perl Programming Language (version 5.26 for Unix) | Perl www.perl.org |
RRID:SCR_018313 |
| Strawberry Perl version 5.24.0.1 (for Windows) | Strawberry Perl Strawberryperl.com |
RRID:SCR_018313 |
| The world-wide-web library for Perl, module 6.39 | LWP Meta CPAN https://metacpan.org/pod/LWP |
N/A |
| Comprehensive Perl Archive Network | https://metacpan.org/ | RRID:SCR_007253 |
| Python version 3.3 (for Unix) | Python www.python.org |
RRID:SCR_008394 |
| Generalized Algebraic Modeling System (GAMS) version 24.7.4 | GAMS Products and Downloads www.gams.com/products/buy-gams/ |
RRID:SCR_018312 |
| CPLEX solver version 12.6 | GAMS Products and Downloads www.gams.com/products/buy-gams/ |
N/A |
| Other | ||
| Holland Computing Center: Crane Computing Cluster (64 GB RAM, Intel Xenon E5-2670 2.60 GHz processor, 2 CPUs per node) | Holland Computing Center https://hcc.unl.edu/ |
N/A |
| Dell Inspiron 7373 model laptop computer (8 GB RAM, Intel Core i5 1.60 GHz processor, and 250 GB solid-state hard drive) | This particular model is no longer in production, but any reasonably up-to-date computer will work | N/A |
Materials and Equipment
Throughout this work, a Dell Inspiron 7373 model laptop computer using the Microsoft Windows 10 Home operating system was used. This computer has a 250 GB solid-state hard drive, an Intel Core i5-8250U CPU @ 1.60 GHz (1,800 Mhz, 4 cores and 8 logic processors), and 8 GB Random Access Memory (RAM). Uses of this computer include directly running program codes which used internet resources and for Secure Shell (SSH) access to the Crane computing cluster at the Holland Computing Center (HCC) of the University of Nebraska – Lincoln. At the time of writing, the Crane cluster is the most powerful cluster at the HCC. The Crane computing cluster has 64 GB of RAM, utilizes an Intel Xenon E5-2670 2.60 GHz processor with 2 CPUs per node. The Crane computing cluster is used primarily for running GAMS codes and for running codes which creates necessary inputs for GAMS codes. It should be noted that the Crane computing cluster is used to for running GAMS code as these codes are used to solve large linear algebra problems with matrices in the order of thousands in both dimensions. Such computationally expensive problems are either very straining to personal computer or even impossible to perform in a reasonable amount of time. Therefore, it is highly suggested that followers of this protocol have access to some advanced computing resource.
Alternatives: For running all code except for that which uses the GAMS programming language, any reasonably up-to-date computer may be used to run the code whether desktop or laptop, and whatever operating system used whether Windows, Mac OS, or Unix/Linux. In the “Before You Begin” section is detailed some software tools which may be used, and indeed are used by other members of the research laboratory to which the authors belong.
For running GAMS software, often access to supercomputing resources are preferable for considerations of time. High research institutions (R1 and R2 institutions by the Carnegie Classification of Institutions of Higher Education) often have either supercomputing facilities of their own or collaborations with other institutions which give them access to supercomputing facilities. For those without such resources, the United State of America, specifically the National Aeronautics and Space Administration (NASA) and the Department of Energy (DOE) have supercomputing facilities which may be used by researchers. The former has several CPUs as part of their High-End Computing Capability (HECC) resource. Use of these resources can be requested by researchers by submitting a request. DOE supercomputing resources are through the supercomputing facilities at the Oak Ridge National Laboratory (ORNL), allows for researchers to apply to use their available supercomputing resources such as Summit, Rhea, and HPSS. The ORNL is home to systems biology research such as the research described in this protocol, including the US DOE Systems Biology Knowledgebase (KBase available at kbase.us).
Step-By-Step Method Details
Iterative Application of OptFill to the Exophiala dermatitidis Draft Models to Development of the Final Model
Timing: days to weeks
Now that the draft model is in a state such that OptFill can be applied, this section details the iterative application of OptFill to the second draft model. This section specifically discusses the applications of OptFill to the second, third, and fourth drafts of the iEde2091 model to expand the functionality of the reconstructions. Detailed in this section is the construction of each database, the application of OptFill, the inclusion of a chosen solution to the previous draft model to produce the next draft model, and the application of BLASTp using NCBI’s BLAST API to determine the evidence for the selected OptFill solution. This method is used in Schroeder et al. (2020) in model reconstruction.
-
1.
Building the first database. From the analysis of the overlap of enzymes present in the four Apsergillus models, the set of enzymes which is common to any three of those models and absent from the fourth should be used as the basis of the first database (from analysis done in Before You Begin step 17). These enzymes should be allowed with all compartmentalizations which are already present in the second draft genome-scale Exophiala dermatitidis model. This list of enzymes with appended compartmentalization (such as “1.1.1.1[c]” for alcohol dehydrogenase in the cytosol) can then be converted to a list of reactions with stoichiometry using the “Enzymes_to_rxns.pl” Perl language code (yellow arrow in Figure 2) which automatically adds compartmentalization to each reaction stoichiometry based on the compartmentalization of the enzyme. The list of reactions and stoichiometries produced by this step is defined as the first database for the application of OptFill.
-
2.
Applying OptFill for the first time. Using the database defined in step 1, OptFill should then be applied to the second draft model of Exophiala dermatitidis. The process of the application of OptFill is the same as that described in the “General steps on how to apply OptFill” section of this protocol, and therefore will not be repeated here (Figure 5 and Figure 6 are taken from the results of this application of OptFill as a method for checking results obtained). OptFill produces two types of results, first the results of the TIC-Finding Problem (TFP) and second, the results of the Connecting Problems (CPs) which provide sets of reactions to be added to the model to fill metabolic gaps. See the “General steps on how to apply OptFill” section of this protocol for how to read and interpret OptFill results.
-
3.
Selecting and applying the first filling solution. In this instance, it is recommended to select and implement the first CPs solution produced by OptFill as this solution improves the model yet not at the expense of adding unnecessary additional reactions. The stoichiometries for the reactions selected by the first CPs solution (taken from the first database file) should be added to a copy of the second draft Exophiala dermatitidis model in order to make the third draft E. dermatitidis model.
Note: Other solutions of the CPs could be selected at this stage but would result in a different model and may result in different results of analyses than those obtained through the iEde2091 model.
-
4.
Performing BLASTp to support the first solution. From the OptFill solution selected, a list of enzymes which catalyze these reactions should be manually gathered from the KEGG database and placed into a list in the file “EClist_1.txt”. Using this list and the code “BidirectionalBLAST.pl” (input files “EClist_1.txt” and “BlastSpecs.txt”) an automated search should be made to determine if there is genetic evidence to support this OptFill solution using the “BidirectionalBLAST.pl” code. This code uses an input list of enzymes; specifications as to acceptable cutoffs values for percent positive substitution of residues and expect value (E value) related to sequence similarity; and a list of related species from which to take the search sequences for the given enzyme. This code works as follows. First, for each enzyme on the list, this code searches KEGG for amino acid sequences for genes known to produce that enzyme from an acceptably related organism (these organisms are specified in the “BlastSpecs.txt” file). This sequence is then used as the query in a BLASTp search, utilizing the BLAST Application Programming Interface (API), against only the Exophiala dermatitidis genome in the NCBI database. It should be noted that the performance of the BLASTp analyses themselves are based on the sample Perl code provided by NCBI for using the BLAST API, available in the developer information of the BLAST API documentation (BLAST Developer Information, n.d.). The time need to receive BLAST results depends not on the BLAST code, but rather the volume of demand for the BLAST tool at the time. All matches are then exported to various text files to store these “forward” BLAST results. This code then evaluates each match in terms of the cutoffs provided, and if the match passes, performs a BLASTp of the sequence found in the target genome against the sequence provided by the reference genome. Should this “backward” BLAST also pass the provide criteria, the match is accepted. The recommended cutoffs could be 60% positive substitution (e.g., 60% of residues are conserved or substituted by a similar amino acid) and an expect value of 1E-30. This should result in a CSV file which details genome-based support for the the inclusion of the reactions included in the OptFill results.
CRITICAL: Note the formatting of input files “BlastSpecs.txt” and “EClist_1.txt”, provided in the GitHub associated with Schroeder et al. (2020), as no effort was made to allow for different formatting of input files in the “BidirectionalBLAST.pl” code.
CRITICAL: A loss of internet connection while the “BidirectionalBLAST.pl” code is running could cause the program to terminate prematurely.
Note: If the “BidirectionalBLAST.pl” code no longer functions, consult the BLAST Developer information web page or other BLAST API documentation to determine if the API has changed or been updated.
Note: It is suggested by the authors to run the “BidirectionalBLAST.pl” code such that times of peak BLAST usage are avoided (or minimized). In the experience of the authors, it is best to run such code overnight or over the weekend.
Note: Depending on how the supercomputing facilities which the reader uses works, this code may need to be run on a personal or work device (such as a laptop or desktop computer) rather than the supercomputing cluster as operations which directly interact with outside websites may not be allowed.
-
5.
Building the second database. The second database should be constructed in the same manner as the first database (detailed in step 1); however, the enzymes used for the construction of this database are those common to two of four of the Aspergillus models analyzed, allowing all compartmentalizations currently in the third draft E. dermatitidis model for each enzyme.
-
6.
Applying OptFill for the second time. OptFill should then be applied to the third draft Exophiala dermatitidis model (serving as the model) with the second database built in step 5 serving as the database for this application of OptFill, applying the procedure detailed in “General steps on how to apply OptFill”. The results of this application are described in Schroeder et al. (2020).
-
7.
Selecting and applying the second filling solution. As with step 3, the best solutions should be selected from the second application of OptFill, and the stoichiometries of the reactions in the optimal CPs solution should be added to a copy of the third draft Exophiala dermatitidis model to produce the fourth draft E. dermatitidis model.
-
8.
Performing BLASTp to support the second solution. As in step 4 a list of enzymes which catalyze the reactions which participate in the OptFill solution selected in step 7 is created, and the “BidirectionalBLAST.pl” code is applied to this list to attempt to find some genomic support for this second OptFill solution.
-
9.
Building the third database. The third database is built in the same manner as the first and second databases (detailed in steps 1 and 5); however, the enzymes which should be used for the construction of the database are those unique to one of the Aspergillus models analyzed, allowing all compartmentalizations currently in the fourth draft E. dermatitidis model for each of these enzymes.
-
10.
Applying OptFill for the third time. As in steps 2 and 6, OptFill should then be applied for the third time using the fourth draft E. dermatitidis model as the model file, the third database (constructed in step 9) as the database, and following the procedure outlined in the “General steps on how to apply OptFill” section of this protocol. The results of this application are described in Schroeder et al. (2020).
-
11.
Selecting and applying the third filling solution. As with steps 3 and 7, the best solutions should be selected from the third application of OptFill, and the stoichiometries of the reactions in the optimal CPs solution can then be added to a copy of the fourth draft Exophiala dermatitidis model to produce the fifth draft E. dermatitidis model.
-
12.
Performing BLASTp to support the third solution. As in steps 4 and 8, a list of enzymes which catalyze the reactions which participate in the OptFill solution of step 11 should be created, and the “BidirectionalBLAST.pl” code is applied to this list to find some genomic support for this third OptFill solution.
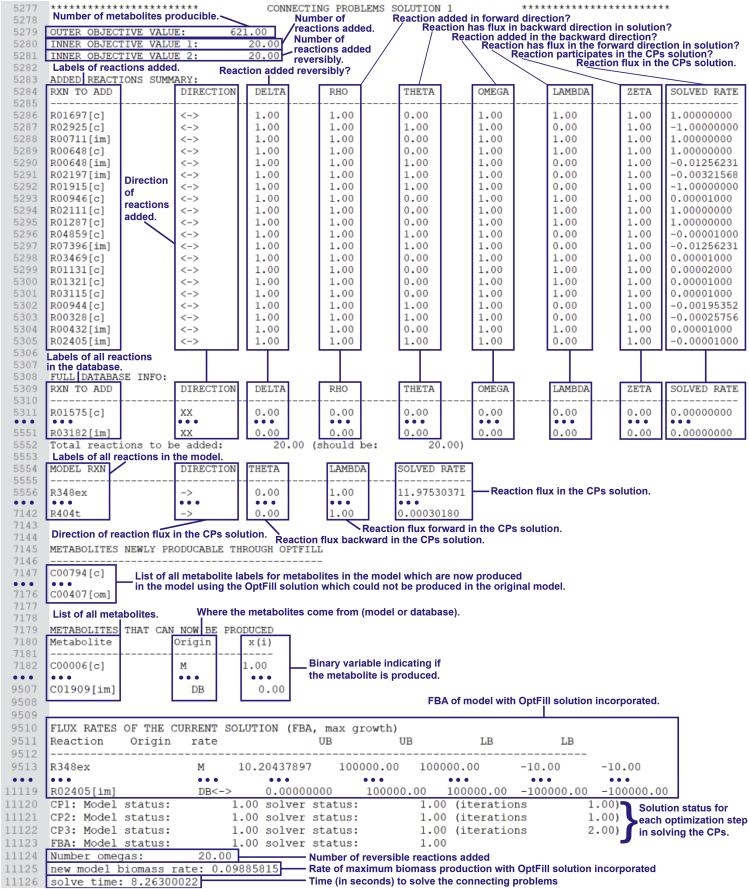
Figure 5.
Screenshot of an Example of the Output of the Standard TIC-Finding Problem (TFP) from the First Application of OptFill to the Second Draft Exophiala dermatitidis Model
Black text is the output, blue text, lines, and boxes serve to highlight important features of the output so that it may be better understood.
Figure 6.
Screenshot of an Example of the Output of the Standard CPs from the First Application of OptFill to the Second Draft Exophiala dermatitidis Model
Black text is the output, blue text, lines, and boxes serve to highlight important features of the output so that it may be better understood.
iEde2091 Model: Shadow Price Analyses
Timing: hours to days
After reconstructing the Exophiala dermatitidis model iEde2091, it is desired to use this model in analysis of E. dermatitidis metabolism. As the defensive pigments of E. dermatitidis are of particular interest for their ability to confer polyextremotolerant properties to an organism, shadow price analysis is selected as a tool for metabolic investigation of these molecules. Shadow price represents the cost to the objective (growth in this model) to produce one more unit (here mmol/gDW∙h) of a particular metabolite. The analysis is performed using the dual form of the FBA optimization problem to determine the shadow price for each metabolite. It can then be used to determine the cost of particular metabolic phenotypes to the organism (here melanogenesis and carotenogensis) to give greater insight into an organism’s metabolism. The shadow price analysis of metabolites, specifically defensive pigments, is one of the key analyses of Schroeder et al. (2020). Here is a step-by-step description of how the shadow price analysis was performed on the fifth draft Exophiala dermatitidis model, which could hopefully aid others in their uses of shadow price analysis. Further, this analysis resulted in some additional curation of the fifth draft Exophiala dermatitidis model, which finally resulted in the iEde2091 model.
-
13.
Get all requisite files. Files which are required for the shadow price analysis are included in the GitHub associated with Schroeder et al. (2020) (doi:10.5281/zenodo.3608172) and indicated by light blue-colored arrows in Figure 2. These files include “iEde2091.txt” (the model file), “convertModel.py” (the file which creates necessary input files from the model file), and “get_shadow_prices.gms” (the file which performs the shadow price analyses).
Note: So as not to upload many duplicates of the “convert∗.py” (where “∗” represents some descriptive word like “Model” or “Database”), the “convertModel.py” code contains a generic placeholder “Model.txt” as the input file in line 20. For this subprocedure, replace this text with “iEde2091.txt”.
-
14.
Setting up appropriate file architecture. For this protocol, it is assumed that all three files mentioned in step 13 are in the same file folder.
-
15.
Run code generating requisite input files for shadow price analysis. For Windows users, open the command prompt and for Mac and Unix users, open the terminal. Navigate the working directory to where the files are located from all previous steps (the iEde2091 should be present in this direcotyr). To run the code, simply type “python convertModel.py >output.txt” into the terminal/prompt followed by selecting enter on the keyboard. The “>output.txt” portion should place what would normally be written to the command prompt or terminal to a text file named “output.txt” for later viewing.
CRITICAL: The format of the model file, here “iEde2091.txt”, is important to how the convert file reads the model and the proper creation of the input files required for the shadow price analysis. The format is as follow (Perl- and python- format regular expressions are used to describe the format, items which will be described in greater detail are bracketed by “<>”):
| “<ReactionLabel>\t(<#>\s<MetaboliteLabel>(\s\+\s)?)∗(\->|<\->|<\-)\s |
| (<#>\s<MetaboliteLabel>(\s\+\s)?)∗” |
-
a.
Example reactions and reaction formats are shown in Figure 7 with the format described below.
-
b.
“<ReactionLabel>”: label of the reaction. While it can be anything, but in this work the reaction label is composed of the KEGG reaction identifier, if applicable, followed by square brackets surrounding a short code indicating subcellular compartment. Exchange, transport, and reactions not in the KEGG database but in the Exophiala dermatitidis metabolism generally have custom reaction labels, which uses three digits after the character “R” (such as “R900[c]” and “R901[e]” in Figure 7), as opposed to five digits in the KEGG identifiers.
-
c.
“<#>”: stoichiometric coefficient of the metabolite in the given reaction. Not to be confused with a line beginning with “#” which denotes a comment in the model file (such as the first line of text in Figure 7). These comments are ignored by the python code “convert∗.py” so that the comments aid in the organization of the file yet have no effect on the actual function of the model.
-
d.
“<Metabolite label>”: label of a metabolite. While it can be anything, in this work the metabolite label is composed of the KEGG reaction identifier, if applicable, followed by square brackets surrounding a short code indicating subcellular compartment. Metabolites which do not have KEGG identifiers have custom labels beginning with “X”, such as X00001[c] in Figure 7 which stands in for 1,8-dyhidroxypaphthalene which is not a compound in the KEGG database.
Note: Any changes made to the model, such as curation or the addition of reactions, should be made to the “iEde2091.txt” model file, then repeating this step (specifically running the “convertModel.py” code) will automatically update all input files required for the shadow price analysis.
-
16.
Run shadow price analysis code. Run the “get_shadow_prices.gms” code using the command “gams get_shadow_prices.gms”. The mathematics related to this analyses are described in detail in Schroeder et al. (2020); however, it may be summarized that the shadow price is the cost to the objective function (in this case, rate of biomass production) that would be incurred by producing one more mmol/gDW·h of a given metabolite. This creates several output files. First is “rxn_rates_out.csv”, which is a CSV file which stores the reaction flux (in mmol/gDW·h) for each FBA performed in the shadow price analysis. The second output file is “shadow_price.csv”, which stores the shadow prices calculated for metabolites which are metabolically close to carotenoids and melanins. The next file is “shadow_price_MCoA.csv” which stores the calculated shadow prices for malonyl-CoA and its precursor metabolites. Finally, “shadow_price_biomass.csv” stores the shadow price of all biomass precursor for each FBA analysis.
-
17.
Study shadow price analysis results. The majority of shadow price values should be negative, indicating that producing extra of any metabolite detracts from biomass production, except for metabolites which may be biomass-coupled. A discussion on biomass-coupling can be found in Burgard, Pharkya and Maranas (2003). This study is generally the most time-consuming step of the procedure of shadow price analysis applied to the iEde2091 model.
-
18.
Model curation using shadow price analysis. When applying shadow price analysis to the fifth draft Exophiala dermatitidis model, it may be the case that some compounds have positive shadow prices (indicating that if more is made then more biomass is also made) and others may have shadow prices which vary between the high, medium, and low limiting nutrient availability conditions. These issues are likely indicators of mass or charge imbalance is some reaction involving that particular metabolite, or a metabolite upstream of that metabolite in the reaction network, or that a particular metabolite is coupled with biomass production. The latter is unlikely unless the model is of a particular strain designed to have biomass production be coupled with metabolite production. The former can be corrected by manual curation which addresses reaction balances. The shadow price analysis may then be run again until neither of these indicators remain present in the analysis. In Schroeder et al. (2020), this served as the final curation step to turn the fifth draft Exophiala dermatitidis model into the final iEde2091 model.
Figure 7.
Screenshot from the “iEde2091.txt” Model Files Showing an Example of the Formatting of the iEde2091 Model
The first line shows an example comment inside the model (comments start with a pound sign “#”). The formatting is such that the reaction label is written (for instance “R10965[c]”), followed by a tab, followed by the stoichiometry of the reaction. Forward (reaction “R02907[c]”), backward (reaction “R02906[c]”) and reversible (reaction “R10965[c]”) reactions are shown, where the direction of the reaction is written into the stoichiometry of the model by the format of the arrow. Further, custom reaction labels (such as “R900[c]” and “R901[c]”) and metabolite labels (“X00001[e]” and “C17937DHN[e]”) are shown in this example. The latter custom label is made from the KEGG identifier for melanin (C17937) combined with a label for the type of melanin (“DHN”) and the metabolite compartmentalization (“[e]”).
General Steps on How to Apply OptFill
Timing: minutes to 1 week
Should a reader wish to apply OptFill to a Genome-Scale Model (GSM) of an organism of choice, as opposed to the iEde2091 model as described in this workflow, this section will describe, in more general terms, the step-by-step procedure to apply OptFill using GAMS. This section describes the required files, directory structure, and modifications to code provided in the GitHub repositories related to this work which will be necessary to apply OptFill to an organism and model of the user’s choice.
-
19.
Getting all requisite files. For the general application of OptFill, several input files are required, and the number of required files varies to some extent by the procedure used. Here it will be assumed that a similar procedure is used in the general application of OptFill as in the specific applications of OptFill to build the iEde2091 model. Therefore, required files include the following (marked by dark blue arrows in Figure 2):
-
a.
One model file – the model whose metabolic gaps are to be filled using the OptFill method (in this case the second draft Exophiala dermatitidis model is marked).
-
b.
One database file – the database of reactions representing metabolic functionalities to use in the filling of metabolic gaps of the model using the OptFill method (the “3of4DB.txt” file is marked as this is what was used with the second draft Exophiala dermatitidis model).
CRITICAL: These files should have the same formatting as described in step 15.
-
c.
Two “convert∗.py” files – Line 20 of each of these “convert∗.py” files should be changed so that one file converts the provided model file, while the other converts the provided database file. Further, lines 23 through 32 in these files must be somewhat changed so that unique sets of output files are made for each file to which as “convert∗.py” code is applied.
-
d.
One “prep_for_optfill.pl” file – This file runs both of the “convert∗.py” and creates certain input files for the OptFill code, such as the set of all metabolites and the set of all reactions in both the model and database.
CRITICAL: Attention should be paid to updating the input files in the “convert∗.py” and “prep_for_optfill.pl” code based on what the codes in this step were named (specifically lines 9 and 10 of this code) and what the output files of step 15 were named (specifically lines 15, 22, and 98 should reference the output files of the convert code which converted the model file, and lines 28, 34, and 103 should reference the output files of convert code which converted the database file).
-
e.
One OptFill file – The OptFill code file which will fill the metabolic gaps in the model file using the database file.
CRITICAL: Attention should be paid to updating the input files in the code based on the name of output files of previous steps, specifically step 15. Lines particularly important to examine are 31, 34, 37, 40, 43, 46, 49, 58, 61, 64, 67, 79, 82, and 85.
-
20.
Setting up appropriate file architecture. These codes assumed that all other codes are contained in the same directory.
Note: The authors set up the directory on the Crane computing cluster of the Holland Computing Center which was used in this work.
-
21.
Run code generating requisite input files for OptFill. This code can be run in two steps: first run the “prep_for_optfill.pl” code (command is “perl prep_for_optfill.pl” provided the terminal working directory is the directory in step 20).
CRITICAL: Format the model and database files (format shown in Figure 7); otherwise code will not run properly.
Note: This step will create a large number of files.
-
22.
Run OptFill. OptFill should be run on the hardware as advance as possible which allows for a long runtime. Depending on the quality and size of the model and database used, the runtime may be between a few seconds and several days.
Note: In both the Schroeder and Saha (2020) and Schroeder et al. (2020) works, the Crane computing cluster was used. In order to have an uninterrupted run time, jobs were submitted using the SLURM research manager which is used by all Holland Computing Center computing clusters.
Note: A discussion about the needed quality of the model and how quality influence runtime can be found in Schroeder and Saha (2020). Should the model itself has a large number of inherent TICs, see the general steps on how to apply the TFP. This can greatly increase the runtime, and the TFP may need to be coupled with manual curation to address model quality issues.
-
23.
Understanding OptFill solutions part 1: the TFP. An example of results for the TFP is shown in Figure 5 and will be used to show what a typical output would look like (important features highlighted in blue). The substeps below describe feature of the output of the TFP.
-
a.
Lines 1 through 5 represent the standard output header, which is always at the top of the output file.
-
b.
Lines 6 through 9 is the standard output block which the TIC-Finding Problem (TFP) returns when there are no more TICs of a given size (where the size in indicated by the value of phi) left to find. In this case, there are no TICs of size 1 (which only occur when there are duplicate reactions in the database and model) or of size 2.
-
c.
Lines 18 through 28 and 31 through 41 show two examples of the information provided by the TFP once a TIC is found.
-
d.
On lines 18 and 31, it is shown that each new TIC is assigned a new, whole number, label.
-
e.
On lines 19–20 and 32–33, the objective value of the number of reactions in the TIC are both reported. As discussed in Schroeder and Saha (2020), the objective function is the minimization of the number of reactions participating in the TIC, and that this number is also fixed by the value of phi, rendering the objective function moot, yet one is still required for optimization. Each pair of objective function and number of reaction pairs should have the same value. If they do not, this indicates that the relaxations allowed by the solver may need to be tightened.
-
f.
Lines 21–22 and 34–35 serve as headers to make the output more human-readable and indicates the start of the table which describes the TIC found in detail.
-
g.
At the start of each line in the table is the reaction label as provided in the model file.
-
h.
The next column in the table is an arrow indicating the direction of flux through the reaction when the reaction is participating in the TIC. Each reaction is allowed to proceed either forward or backward. The direction of the reaction is also indicated by the binary variables alpha and beta reported in the last two columns where alpha indicates forward while beta indicates backward.
-
i.
The third column of the table indicates where the reaction resides, either in the model (M) or database (DB).
-
j.
The fourth column reports on the binary variable eta, which simply reports whether (value 1) or not (value 0) a reaction is participating in the found TIC. All values of this column should be 1, and if this is not the case, consider tightening the relaxations allowed by the solver.
-
k.
The fifth column indicates the flux through the reactions participating in the TIC. The values will always belong to the set . This is because allowing smaller magnitude flux rates or more orders of magnitude for the range in flux rates may cause issues with the relaxations used by optimization problem solvers. Generally, the magnitude of the reported flux rate is unimportant, rather the ratios between flux rates is more enlightening. See Schroeder and Saha (2020) for a discussion on this issues.
-
24.
Understanding OptFill solutions part 2: the CPs. An example of the results which may be returned from the CPs is shown in Figure 6. Note that as there are several output sections for each CPs solution, only one example solution is shown, and some lines have been removed from that solution (replaced with ellipses) for the brevity of the figure. It should be noted that, for some portions of the output of the solutions to the CPs, formatting is not as neat as that of the TIC-Finding Problem.
-
a.
Line 2,577 is the heading for the start of the current CPs solution.
-
b.
Lines 5,279, 5280, and 5,281 report the objective solution for the first, second, and third CPs respectively. These problems sought to maximize the number of metabolites which can be produced, minimize the number of reactions, and maximize the number of reactions added reversibly respectively.
-
c.
Lines 5,283 through 5,305 is the first result table of the CPs solution and summarizes which reactions are added in the solution and how those reactions are added. The meaning of each column is detailed in Figure 6.
-
d.
Lines 5,308 through 7,142 is the same table as lines 5,283 through 5,305, though these lines detail the results for all reactions contained in the database, hence the much larger size. This is mostly used for the purposes of debugging. Reactions not included in the CPs solution are labeled with a direction of “XX”.
-
e.
Lines 7,145 to 1,776 list the metabolites which can now be produced by the OptFilled model which the model could not previously produce.
-
f.
Lines 7,179 through 9,507 produces a table of all metabolites in both the model and the database, and the binary variable, x(i), which determines whether or not they are produced.
-
g.
Lines 9,510 through 11,119 show the results of a FBA of the OptFilled model. The alignment of text is not as high quality as with other tables produced by OptFill, but this can be remedied by copying-and-pasting this table into a Microsoft Excel worksheet and using the text import wizard tool to delimit the cells by spaces. This will result in a more readable table of FBA results. This table is mostly used for the purposes of debugging.
-
h.
Lines 11,121 through 11,123 report the model and solver statuses of the solutions, as well as the number of iterations needed for the solver to reach these solutions. Should the model or solver statuses not have a value of 1 at any stage, this would be an indication of a need for debugging the code.
-
i.
Line 11,124 again states the number of reversible reactions in the CPs solution.
-
j.
Line 11,125 states the growth/biomass rate of the OptFilled model, as often fixing metabolic gaps can have an effect on the rate of biomass production.
-
k.
Line 11,126 finally reports the time, in seconds, which the solver takes to reach this optimal solution.
-
25.
Select Solution. A set of unique filling solutions will be provided by the OptFill code. Users should carefully review the filling solutions produced by the OptFill code and select the solution based on criteria such as how optimal the solution is determined by OptFill (e.g., order of solutions), which metabolites are fixed (literature evidence for fixed metabolites), which reactions are added (literature evidence for metabolic functions), or other user-defined criteria.
General Steps on How to Use the TIC-Finding Problem of OptFill for Identifying Inherent TICs
Timing: minutes to 1 week
This section describes how to apply the modified TFP of OptFill to identify inherent TICs utilizing GAMS. This is useful in order to identify TICs inherent to a model to improve the quality of the reconstruction. This section describes the required files, directory structure, and modifications to code provided in the GitHub repositories related to this work which will be necessary to apply the modified TFP to an organism and model of the user’s choice.
-
26.
Getting all requisite files. For the general application of the TFP of OptFill, a few input files are required, and the number of required files varies to some extent by the procedure used. Here it will be assumed that a similar procedure is used in the general application of OptFill as in the specific applications of OptFill to build the iEde2091 model. Therefore, required files include the following (marked by orange arrows in Figure 8):
Figure 8.
Screenshot of a Sub-folder of the OptFill GitHub Repository (https://doi.org/10.5281/zenodo.3518501 or https://github.com/ssbio/OptFill) with code highlighted which can be used to perform the TIC-Finding Problem only on the iJR904 model to identify TICs inherent to the model. This code should be adaptable to other genome-scale models as well.
-
a.
One model file – the model to which the TFP will be applied. Here, the iJR904 model file (“iJR904.txt”) is marked as the system to which the inherent TIC-Finding will be applied.
CRITICAL: These files should have the same formatting as described in step 15.
-
b.
One “convert∗.py” file – A Python code which makes several input files necessary for the TFP. Line 20 of this “convert∗.py” file should be changed so that one file converts the provided model file. (Here, the file is “convert_iJR904.py”).
-
c.
One TIC-Finding Problem file – The code which performs the TIC-Finding Problem on the input model file (here labeled “OptFill_TFP_only_iJR904.gms”).
-
27.
Setting up appropriate file architecture. These codes assumed that all other codes are contained in the same directory.
-
28.
Run code generating requisite input files for OptFill. The “convert∗.py” code from step 26b should be run to create all necessary input files.
-
29.
Run TFP Code. The TFP should be run on the hardware as advance as possible which allows for a long runtime. Depending on the quality and size of the model and database used, the runtime may be between a few seconds and several days.
Note: In both the Schroeder and Saha (2020) and Schroeder et al. (2020) works, the Crane computing cluster was used. In order to have an uninterrupted run time, jobs were submitted using the SLURM research manager which is used by all Holland Computing Center clusters.
-
30.
Analyze Solution. TICs inherent to the model will be included in the solution of the TFP code. These solutions will include both the reactions participating in TICs and their directions in the participating TIC. Inherent TICs generally need to be addressed through manual curation that involve changing reaction directions or removing reactions.
General Steps on How to Use the iEde2091 Model Including Accompanying Codes for Basic Analyses (FBA and FVA)
Timing: seconds to minutes
This section describes how to use the iEde2091 model for general analyses such as FBA and FVA using codes provided by the authors. The code focused on in this section utilize GAMS.
-
31.
Getting all requisite files. To run FBA and FVA codes on the iEde2091 code, four files are required (indicated by purple arrows in Figure 2).
-
a.
“iEde2091.txt” file – The model file for the iEde2091 model.
-
b.
“convertModel.py” – Converts the iEde2091 model file into several files required by FBA and FVA code.
-
c.
“FBA.gms” file – Code which runs Flux Balance Analysis on the iEde2091 model.
-
d.
“FVA.gms” file – Code which runs Flux Variability Analysis on the iEde2091 model.
-
32.
Set up appropriate file architecture. These codes assume that all files are contained in the same directory.
-
33.
Run code which generates input files necessary for FBA and FVA. The “convertModel.py” file should be run to create all requisite input files (the command is “python convertWLS.py”).
-
34.
Run FBA and FVA. Flux Balance and Flux Variability Analyses may be run using the command “gams F∗A.gms” (where “∗” is substituted with “B” or “V” based on the desired analysis), and each will create output files with the results of these analyses.
iEde2091 Model: Comparison with Human Metabolism
Timing: minutes to days
As mentioned in (Schroeder et al., 2020), E. dermatitidis is a potential model defensive pigment producing organism due to it relatively small genome of 26.4 Mbp, compared to other model organisms including Saccharomyces cerevisea with a genome of 11.9 Mbp, Drosophilia melanogaster with a genome of 137.6 Mbp, Arabidopsis thaliana with a genome of 119.1 Mbp, and Homo sapiens with a genome of 2,893.9 Mbp (National Center for Biotechnology Information, n.d.). Due to the size of the Exophiala dermatitidis genome and the importance of melanin to the organism, the similarities and differences between E. dermatitidis and H. sapiens was investigated to determine the potential of E. dermatitidis as a model of H. sapiens melanogenesis. This section describes the procedure used to compare human and Exophiala dermatitidis melanin metabolisms in the Schroeder et al. (2020). This procedure includes comparing metabolic pathways and comparing human and E. dermatitidis tyrosinase amino acid sequences.
-
35.
Comparing metabolic pathways of melanin synthesis. From the reconstruction of the iEde2091 model, the metabolic pathway of melanin synthesis should be known through the reconstruction process. Now the metabolic pathways for the synthesis of melanins, specifically pheomelanin and eumelanin, must be investigated. Several sources were identified in Schroeder et al. (2020) which detailed eumelanin and pheomelanin synthesis pathways. This allows for a comparison of reaction pathways such as is shown in Figure 3A of Schroeder et al. (2020).
-
36.
Ensuring that all tyrosinase enzymes have been identified in Exophiala dermatitidis. The key enzyme in the synthesis of eumelanin (and also the synthesis of pheomelanin in humans) is tyrosinase (EC number 1.14.18.1). In Chen et al. (2014), this four gene copies encoding the tyrosinase enzyme are listed with NCBI accessions of XP_009160170.1, XP_009156893.1, XP_009157733.1, and XP_009155657.1. The former two are said to be gene copies unique to E. dermatitidis, whereas the latter two are said to be conserved from Aspergillus homologs. A quick check should be done to ensure that no new tyrosinase gene copies have been identified.
-
a.
Perform a search in NCBI using the string “tyrosinase Exophiala dermatitidis”, which should produce a short list of all enzymes annotated as tyrosinase in Exophiala dermatitidis. In Schroeder et al. (2020), these were the only four gene copies in the list.
-
b.
Perform a BLASTp search to determine if there are as yet unidentified tyrosinase gene copies in the Exophiala dermatitidis genome. Do this by performing a BLASTp of the known E. dermatitidis gene copies against the E. dermatitidis genome. This can be done by searching for the accession number in NCBI and selecting the protein page result matching that accession number. Then, selecting the “FASTA” link on that page (just below the heading). This will give the amino acid sequence of the tyrosinase gene copy.
-
c.
The amino acid sequence can then be copied-and-pasted into the query sequence textbox in the the BLASTp tool (see Figure 9). Figure 9 shows the BLASTp settings used. Figure 10 shows composite results of the BLAST results performed on 04/23/2020. From this, it can be shown that, at present, there is no genetic evidence for additional gene copies of tyrosinase in E. dermatitidis using this method. Interestingly, some tyrosinase genes are so dissimilar, at least on the whole amino acid sequence, that some gene copies do not match to all other gene copies of tyrosinase.
Figure 9.
Setup of the BLASTp Query Used to Map Exophiala dermatitidis Tyrosinase Gene Copy XP_0099155657.1 Back onto the Exophiala dermatitidis Genome to Search for as yet Unidentified Tyrosinase Gene Copies
This search was repeated for each gene copy of Exophiala dermatitidis.
Figure 10.
BLASTp Results for the Query of Each Exophiala dermatitidis Tyrosinase Gene Copy against the Exophiala dermatitidis Genome when Searching for Previously Unidentified Tyrosine Gene Copies
(A) Search results for comparing E. dermatitidis protein XP_009156893.1 (tyrosinase) to the rest of the E. dermatitidis genome utilizing the settings shown in Figure 9.
(B) Search results for comparing E. dermatitidis protein XP_009157733.1 (tyrosinase) to the rest of the E. dermatitidis genome utilizing the settings shown in Figure 9.
(C) Search results for comparing E. dermatitidis protein XP_009160170.1 (tyrosinase) to the rest of the E. dermatitidis genome utilizing the settings shown in Figure 9.
(D) Search results for comparing E. dermatitidis protein XP_009155657.1 (tyrosinase) to the rest of the E. dermatitidis genome utilizing the settings shown in Figure 9.
-
37.
Comparison of human and tyrosinase enzymes through BLASTp. As one method for evaluating the similarity of human and Exophiala dermatitidis melanin synthesis, the similarity between tyrosinase enzymes of the two species should be evaluated. The first method of evaluation is a BLASTp analysis, which can be performed similarly to steps 4, 8, and 12, except that the search is limited to the Homo sapiens genome. The combined results for this search can be seen in Figure 11. In summary, no tyrosinase gene copy from Exophiala dermatitidis has high sequence similarity to that of Homo sapiens.
Figure 11.
BLASTp Result for the Query of Each Exophiala dermatitidis Tyrosinase Gene Copy against the Homo Sapiens Genome to Determine if the Tyrosinase Gene of Humans Are Sufficiently Similar to that of E. dermatitidis to Produce a Significant BLASTp Match
(A) BLASTp results comparing E. dermatitidis protein XP_009156893.1 (tyrosinase) to the human (H. sapiens) genome.
(B) BLASTp results comparing E. dermatitidis protein XP_009157733.1 (tyrosinase) to the human (H. sapiens) genome.
(C) BLASTp results comparing E. dermatitidis protein XP_009160170.1 (tyrosinase) to the human (H. sapiens) genome.
(D) BLASTp results comparing E. dermatitidis protein XP_009155657.1 (tyrosinase) to the human (H. sapiens) genome.
-
38.
Comparing human and tyrosinase enzymes through COBALT. A BLASTp analysis generally looks a whole-sequence similarity; however, the whole amino acid sequence of an enzyme is generally not critical to enzyme function. Rather, those amino acids in the active site or sites of an enzyme are most critical; therefore, this step is focused on evaluating the similarity and conservation of the active site using the Constraint-based Multiple Alignment Tool (COBALT) (Papadopoulos and Agarwala, 2007). The COBALT is available through NCBI (url: https://www.ncbi.nlm.nih.gov/tools/cobalt/re_cobalt.cgi) and is relatively simple to use, as described below to compare human tyrosinase, human tyrosinase related proteins, and Exophiala dermatitidis tyrosinase gene copies.
-
a.
To compare human tyrosinase related proteins, human tyrosinase alleles, and Exophiala determatitidis, use the settings and search query shown in Figure 12 (note that since this screenshot was taken on 04/23/2020, the COVID-19 banner was removed from the image so as to show what is normally seen), then select “Align”.
Figure 12.
Input to the COBALT Tool Used when Comparing the Amino Acid Sequences of Human Tyrosinase-Related Proteins 1 and 2, Three Different Human Alleles for Tyrosinase, and the Four Gene Copies of Tyrosinase in E. dermatitidis
-
b.
Once the alignment is complete, the enzymes may be compared, particularly in term of conserved sequences. For this analysis, 3-bit conservation settings should be used (this can be changed by changing the drop-down menu labeled “Conservation Setting:” under the heading “Alignments”).
-
c.
The active sites of tyrosinase and tyrosinase related proteins, CuA and CuB (standing for Copper-binding domains A and B respectively) can be identified using information on the residue positions of these active sites from sources such as García-Borrón and Solano (2002), Spritz et al. (1997), and Furumura et al. (1998). It should be assumed that the sequences from E. dermatitidis genes copies of tyrosinase which align with these active sites are the active sites of these tyrosinases. In summary, these results should show relatively well-conserved active sites and particularly well-conserved key active site residues.
-
39.
Comparing E. dermatitidis tyrosinase gene copies to hidden Markov Models (hMMs) using Pfam. To further evaluate how similar Exophiala dermatitidis gene copies are to other tyrosinase sequences, the sequence of each gene copy should be evaluated against the hidden Markov Models (hMMs) of various protein families using the Pfam tool. As with the COBALT tool, it is fairly simple to use and the steps are described below.
-
a.
The Pfam tool can be found at pfam.xfam.org. To submit a sequence to query against hMMs of protein families, select the link “Sequence Search” in the middle of the page.
-
b.
This will result in a text box appearing in the middle of the page. Copy-and-past the amino acid sequence of the desired protein into that text box and then select “Go” (note that this should be done one sequence at a time).
-
c.
In general, the only protein family to which Exophiala dermatitidis tyrosinase gene copies map is tyrosinase, showing that the important portions of the sequence are well conserved (expect values of 2E-38 or better). Two of these gene copies, XP_0091601701.1 and XP_009156893.1, also had weak matches to the Tyrosinase C hMM as well. A combination of the results for each tyrosinase gene copy of E. dermatitidis is shown in Figure 13.
Figure 13.
Results of the Analysis of Each Tyrosinase Gene Copy in Exophiala dermatitidis in a Hidden Marko Model Analysis Done by the Pfam Tool
As shown here, each of these sequences align well with the tyrosinase hidden Markov Model.
(A) Result of comparison of E. dermatitidis protein XP_009160170.1 (tyrosinase) against the baseline Hidden Markov Chain of the tyrosinase family using the Pfam tool.
(B) Result of comparison of E. dermatitidis protein XP_009156893.1 (tyrosinase) against the baseline Hidden Markov Chain of the tyrosinase family using the Pfam tool.
(C) Result of comparison of E. dermatitidis protein XP_009157733.1 (tyrosinase) against the baseline Hidden Markov Chain of the tyrosinase family using the Pfam tool.
(D) Result of comparison of E. dermatitidis protein XP_009155657.1 (tyrosinase) against the baseline Hidden Markov Chain of the tyrosinase family using the Pfam tool.
-
40.
Analyze all data gather to this point on the similarity of Human and Exophiala dermatitidis melanin synthesis. To this point, a comparison of the reaction pathway which produces melanins has been made (step 35), the key enzyme (tyrosinase) is compared between the two species (steps 37, 38, and 39), and tyrosinase gene copies of E. dermatitidis are compared to the hidden Markov Models of tyrosinase. At this stage, comparisons of human and Exophiala dermatitidis eumelanin and pheomelanin metabolism may be made. An example of such an analysis is provided in Schroeder et al. (2020).
Expected Outcomes
There are three primary outcomes of this protocol. The first two deal directly with the subject of this protocol, that is Exophiala dermatitidis. First is the iEde2091 model, which is a curated metabolic model of Exophiala dermatitidis. This model can be put to a wide variety of purposes such as hypothesis-driven discovery, metabolic engineering, model-guided discovery, the integration of ‘omics’ data, medical studies (as E. dermatitidis may be a model organism for human melanocytes yet is also an infectious organism), and other purposes to which metabolic models have been put as detailed in works such as Oberhardt, Palsson and Papin (2009); Feist and Palsson (2008); and Zhang and Hua (2016). Code and procedures use and described in this protocol will also make the use of this model easier so that it may be more readily usable to non-experts. The second primary outcome is the analysis of the comparability of human and E. dermatitidis melanin synthesis pathways. This analysis is visually summarized in Figure 14 using pieces of images from Schroeder et al. (2020). The third primary outcome deals with the understudied nature of Exophiala dermatitidis, yet a successful genome-scale metabolic reconstruction of the organism was achieved. Here, it should be clarified as to what a successful reconstruction is in the context of an understudied organism. Due to the nature of understudied organisms and the lack of in vivo experimental procedures included in this protocol, a “successful” model is not defined here as one that closely replicates in vivo behavior because such comparisons may not be able to be made. Rather, a “successful” metabolic model of an understudied organism should be able to: i) simulate growth, ii) make full use of current knowledge of the organism, iii) be able to be subject to in silico metabolic analysis techniques to support, and iv) usability as a hypothesis-generating tool for the design of in vivo experiments to expand knowledge of the understudied organism. The hypotheses generated through this protocol, detailed in Schroeder et al. (2020), are currently under in vivo investigation by a collaborator. This protocol then has the potential to serve as a guideline for other investigators interested in in silico experimentation of other understudied organisms to show how a GSM may be reconstructed.
Figure 14.
Figure Highlighting the Expected Results of This Protocol and Their Relationships while “Black Boxing” Much of the Procedure (Red Arrows)
Beginning with knowledge of the Exophiala dermatitidis system and genome (upper left hand corner) and four published genome-scale models of Aspergillus species, various techniques including OptFill are used to create the first major result: the genome-scale model (GSM) of E. dermatitidis. This model, in conjunction with knowledge of the human genome and the E. dermatitidis genome, was used to evaluate the similarity between human and E. dermatitidis melanogenesis for the second major result of this protocol. The third major result is the study of the shadow prices of defensive pigments, namely carotenoids (pink rods) and melanins (brown triangles), which showed that carotenoids are more expensive, suggesting a heretofore undiscovered role for carotenoids.
Limitations
Automated reconstruction processes for GSM of metabolism exist, including through resources like KBase (Arkin et al., 2018) and ModelSeed (modelseed.org); however, these make use of a different philosophy of model reconstruction than used in this work. These tools seek to create a well-connected metabolic network at the cost of a “permissive” model reconstruction. That is, low confidence reactions will often be added to the model during reconstruction, and few methods exist to identify and address infeasible cycles. Therefore, often the models reconstructed through these methods contain functions which may not be present in the organism and TICs, both of which require manual curation to address. Therefore, these automated methods may be best used as a method to produce draft models, rather than high-quality reconstructions. This protocol, in contrast, outlines a conservative reconstruction process, including a conservative reconstruction tool (OptFill), which seeks to minimize the number of new functionalities added, and avoid TICs, while sacrificing connectivity. This “conservative” reconstruction approach is well-suited for the GSM reconstruction of understudied organisms.
Most of this protocol describes how to recreate the iEde2091 model presented and analyzed in (Schroeder et al., 2020); however, perfect replication of this model following this procedure is likely not possible, as much of the data used in the reconstruction detailed was collected before 2018, yet since then knowledge of E. dermatitidis has increased through more genome assemblies such as shown in step 10 and Figure 3 which acknowledges newer genome assemblies. Further, perfect replication of these works may not be desirable, as no new insights would be gained. Rather, this procedure outlines a method which may be followed for the reconstruction of future E. dermatitidis genome-scale models or genome-scale models of other understudied organisms. This procedure also details how to use the OptFill method, giving greater step-by-step detail of how to use provided code than was possible in Schroeder and Saha (2020) and Schroeder et al. (2020) which are focused on the mathematics of the OptFill method. In general, this protocol should be treated as a procedure to parallel in other research efforts, rather than duplicate.
As noted in the “Before You Begin” section, not all code provided here can be run for free, especially since access to the GAMS programming language requires significant capital investment. The GAMS language leaves users of this protocol with two (at present unequal) options: i) purchase GAMS and the CPLEX solver (the quickest, most tractable, and most computationally powerful option), or ii) adapt the optimization-based tools used throughout the text to some other programming language which is free of charge (such as Python) or already owned (such as MATLAB). To aid in the latter option, please see Schroeder and Saha (2020) and Schroeder et al. (2020) for a detailed mathematical formulation of OptFill which could be adapted to any programming language with optimization capabilities with effort.
To make this protocol as usable as possible by those lacking the resources to purchase GAMS, the authors have attempted to implement OptFill using python and the COBRApy package, the python package for COnstraint-Based Reconstruction and Analysis (COBRA). Upon reviewing the implementation and its performance, the authors have concluded that OptFill in COBRApy is limited by the computational speed of COBRApy. For instance, the COBRApy adaptation of OptFill could not duplicate the first application of OptFill to the draft E. dermatitidis model in a reasonable period of time. It took more than two days to identify the first TIC between the database and model utilizing a supercomputing cluster, suggesting that the full application of the algorithm could have a runtime of months. Therefore, the COBRApy version of OptFill have been implemented and applied to a much smaller problem, the first test model and database from Schroeder and Saha (2020) and is included in the GitHub repository for OptFill (https://doi.org/10.5281/zenodo.3518501). A short protocol detailing how to run OptFill using COBRApy is included in this repository as well. This resulted in total runtimes of 553.4 s to solve the TFP utilizing COBRApy (as opposed to 13.2 s utilizing GAMS/CPLEX) and 737.6 s to solve the Connecting Problems utilizing COBRApy (as opposed to 32.76 s in GAMS). Both results indicated that COBRApy is between one and two order of magnitude slower than GAMS for solving MILP problems such as OptFill for these small test problems. As it was noted in (Schroeder and Saha, 2020), the time to solve OptFill in GAMS increases exponentially as the model and database to which it is applied becomes progressively larger. Should a similar pattern exist in COBRApy, this would explain why the time required to apply OptFill for a model such as iEde2091 would be infeasibly long. In addition to implementing OptFill in python, python-based analyses codes, namely for FBA, FVA, and shadow price analysis, are included in the GitHub repository for iEde2091 in an attempt to make the results of this protocol usable by those without access to GAMS.
Should COBRApy become significantly quicker in solving Mixed Integer Linear Programming (MILP) problems, it may be worth re-investigating the implementation of OptFIll in this package and language in future. Particularly, it appears that COBRApy is significantly slower to identify infeasible problems. For example, when all potential TICs of size 11 between the model and the database have been identified, GAMS concludes that no further solutions exist in 0.08 s, whereas COBRApy requires 20.1 s to make the same determination. Similar four orders of magnitude differences in solution times when determining a problem to be infeasible exist throughout the application of the TFP to the first test model and database. As OptFill often makes use of infeasible results (such as when to advance the size of sought TICs), one of the most promising bottlenecks to address is the quick identification of infeasibility. This can be further evidenced by the fact that, of the 553.4 s needed to solve the TFP, 498.2 s (90% of the runtime) were used while trying to conclude that the model was infeasible. This would not, however, address the runtime disparity between COBRApy- and GAMs- implemented CPs (as infeasibilities are not used in the CPs).
A further limitation of this protocol is that it cannot be entirely automated due to multiple factors. First, steps such as Before You Begin step 16 are difficult to automate as they generally require complex reasoning (such as why to choose one species or group of species over another for model curation and database creation). Second, online tools used in this workflow such as CELLO (in Before You Begin step 14) and BRENDA (in Before You Begin steps 9 and 11) cannot be automated as they lack APIs. When these steps are automated, these automated procedures may be easily undone by the reformatting of the website or tool. Finally, some steps in this protocol require data which is not included in some databases. For instance, KEGG does not contain identifiers for pheomelanin, DHN-melanin, and some melanin precursors, requiring careful manual curation and literature searching for identification and description of the correct melanogenesis synthesis pathways in the iEde2091 model. These tools, and more broadly this protocol, were chosen and used instead of a fully automated model reconstruction procedure such as those available through ModelSeed and KBase because it allows for a more cautious and conservative reconstruction process which is valued given the lack of knowledge of the system and the authors desire not to add more unsupported metabolic functionalities than necessary. Additionally, a previously fully automated process would not have allowed for a demonstration of a new tool to address metabolic gaps such as OptFill.
Troubleshooting
A particularly problematic aspect of this protocol is its use of and reliance on code files. It should be noted that programming codes, especially those reliant on websites or APIs will inevitably not function as described at some point in the future. Basic familiarity with the programming language in question and programming in particular may allow adaptation of codes which no longer work to restore function. In most cases, a “CRITICAL” or “Note” is provided where code is introduced which may be most likely subject to being outdated and some suggestions as to how remedying issues of non-functionality may be possible. Similarly, screenshots taken of websites will also be inevitably outdated at some time in future, through this is probably less critical. For some specific instances of codes which may be more subject to obsolescence, potential solutions are highlighted here.
Problem 1
Major version change(s) in programming languages causing codes included in this protocol to be outdates and not function.
Potential Solution
For those familiar with programming and the synthax of the languages in question, the best solution would be to update the syntax in the code. If this is not possible, then there is a brief description for each coding language as to how backward compatibility may be achieved.
For GAMS: According to GAMS documentation “GAMS makes every attempt to be backwards compatible and ensure that new GAMS systems are able to read save files generated by older GMAS systems.” This being said, GAMS has a Boolean general option called forceWork which can attempt to process files which have an execution error to attempt to be more backwards compatible.
For Python: The version of python used as runtime can be specified from the command line. For instance, “python2 codename.py” (where codename is the name of the code file) will run the code using Python version two, whereas “python3 codename.py” will run the code using using Python version 3. Further, it appears that several version of the Python library can be downloaded (from python.org) and so old releases may be downloaded and used.
For Perl: Version used at runtime can be specified by the command “use VERSION” or “require VERSION” where VERSION is replaced by a string which represent the version which is to be run, for instance “use v5.24.0.1”. This “use” command will get its own line in the Perl code itself.
For all: Another potential solution to this issue is to build an application containerization for each version of each programming language used in this protocol, which allows an application to be self-contained and run a native instance of the programming language it uses. This allows the containerized application to remain viable despite language version changes or updates. This protocol will not utilize application containerization, as the primary objective is to help other be able to utilize a mathematical tool (OptFill) rather than programming tools (other codes used in this protocol).
Problem 2
Two APIs are used in this protocol by various codes in this protocol, the BLAST API and the KEGG API. One potential problem for this protocol in future is the obsolescence of code by updates to these APIs.
Potential Solution
Consult documentation of the API, particularly those documents that are changed.
For BLAST API: Check the provided BLAST Perl code from NCBI to see if there is a need to update the function in common_functions.pl. The code which serves as the basis of the BLAST function used by the “BidirectionaBLAST.pl” code can be found at https://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=DeveloperInfo and downloaded by clicking the “sampler perl code” link. Fixing the BLAST code in particular will require basic programming knowledge and knowledge of the Perl programming language (since it was modified from a stand-alone script to a function). Since all applications interfacing with APIs are written in the Perl programming language, the author would suggest the text “Learning Perl” by Foy, Phoenix, and Schwarz as this text should contain all knowledge necessary to update the code.
For KEGG API: This API is less complex than the BLAST API, and if code interacting with the KEGG API is rendered obsolete, it is likely that the formatting of the URLs for the KEGG API or that the text resulting from those URLs has changed. This can likely be fixed by reviewing and updating calls to the KEGG API used in various Perl languages codes accompanying this work and/or by updating regular expression searches in in the KEGG APIs to address new formatting.
Problem 3
The code which interacts with the BRENDA database no longer works, namely “UniProt_get_ECs.pl” (in Before You Begin step 2) and “NCBI_get_ECs.pl” (in Before You Begin step 4).
Potential Solution
Unfortunately, this is the most susceptible code to obsolescence as BRENDA does not have an API and therefore the code was forced to function by using “screen scraping” techniques. The solution to this problem would likely be to adapt the code to the new format of BRENDA, often by changing the regular expressions used in the code.
Problem 4
This work is quite out-of-date and the time necessary to update it is not considered worthwhile.
Potential Solution
As much of this protocol utilizes interfaces (such as BLAST API and KEGG API), websites (such as BRENDA), and programming languages (GAMS, Perl, and Python) which are subject to change, the authors do not intend this to be a monolithic procedure that can be forever duplicated with the provided code. Rather, in future, it is hoped that this protocol outlines a procedure which may be followed by others for the study of Exophiala dermatitidis or other understudied organism which has shown success with Exophiala dermatitidis. It is also hoped that this protocol will allow for more widespread use of the iEde2091 model, particularly for hypothesis generation related to defensive pigment production, and that it will spur interest in E. dermatitidis as a model organism for human melanocytes.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Technical Contact, Wheaton L. Schroeder (wheaton@huskers.unl.edu) or by the Lead Contact Rajib Saha (rsaha2@unl.edu).
Materials Availability
This study did not generate unique reagents.
Data and Code Availability
The datasets and code generated during this study are available in two GitHub repositories related to this work, one for the Exophiala dermatitidis model iEde2091 (https://doi.org/10.5281/zenodo.3608172) and one for the OptFill tool (https://doi.org/10.5281/zenodo.3518501). These DOIs are for single releases of these GitHub repositories. Repositories which will be updated periodically and can be found through the laboratory group’s GitHub repository pages located at https://github.com/ssbio/E_dermatitidis_model (iEde2091 model and related code) and https://github.com/ssbio/OptFill (OptFill and related code). All data generated in the execution of the protocol has been included in the in appropriate GitHub repository or in the supplemental files provided with the published works associated with this protocol, namely Schroeder and Saha (2020) and Schroeder et al. (2020).
Acknowledgments
This work has been completed utilizing the Holland Computing Center of the University of Nebraska, which receives support from the Nebraska Research Initiative. The authors gratefully acknowledge the APC discount given to them by STAR Protocols which makes this work possible. The authors gratefully acknowledge funding from University of Nebraska – Lincoln (UNL) Faculty Startup Grant 21-1106-4038 as well as the NSF EPSCoR Center for Root and Rhizobiome Innovation Grant 25-1215-0139-025 at UNL.
Declaration of Interests
The authors declare no competing interests.
Author Contributions
Conceptualization, W.L.S. and R.S.; Data Curation, W.L.S.; Formal Analysis, W.L.S.; Funding Acquisition, R.S.; Investigation, W.L.S.; Methodology, W.L.S.; Project Administration, R.S.; Resources, R.S.; Software, W.L.S.; Supervision, R.S.; Validation, W.L.S.; Visualization, W.L.S.; Writing – Original Draft, W.L.S. and R.S.; Writing – Review & Editing, W.L.S. and R.S.
Contributor Information
Wheaton L. Schroeder, Email: wheaton@huskers.unl.edu.
Rajib Saha, Email: rsaha2@unl.edu.
References
- Andersen M.R., Nielsen M.L., Nielsen J. Metabolic model integration of the bibliome, genome, metabolome and reactome of Aspergillus niger. Mol. Syst. Biol. 2008;4:178. doi: 10.1038/msb.2008.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkin A.P., Cottingham R.W., Henry C.S., Harris N.L., Stevens R.L., Maslov S., Dehal P., Ware D., Perez F., Canon S., Sneddon M.W. KBase: The United States department of energy systems biology knowledgebase. Nat. Biotechnol. 2018;36:566–569. doi: 10.1038/nbt.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BLAST Developer Information. (n.d.). National Center for Biotechnology Information. Retrieved April 21, 2020, https://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=DeveloperInfo.
- Burgard A.P., Pharkya P., Maranas C.D. OptKnock: A bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng. 2003;84:647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- Chan S.H.J., Cai J., Wang L., Simons-Senftle M.N., Maranas C.D. Standardizing biomass reactions and ensuring complete mass balance in genome-scale metabolic models. Bioinformatics. 2017;33:3603–3609. doi: 10.1093/bioinformatics/btx453. [DOI] [PubMed] [Google Scholar]
- Chen Z., Martinez D.A., Gujja S., Sykes S.M., Zeng Q., Szaniszlo P.J., Wang Z., Cuomo C.A. Comparative genomic and transcriptomic analysis of Wangiella dermatitidis, a major cause of phaeohyphomycosis and a model black yeast human pathogen. G3 (Bethesda, Md.) 2014;4:561–578. doi: 10.1534/g3.113.009241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R., Chowdhury A., Maranas C.D. Using gene essentiality and synthetic lethality information to correct yeast and CHO cell genome-scale models. Metabolites. 2015;5:536–570. doi: 10.3390/metabo5040536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David H., Özçelik I.Ş., Hofmann G., Nielsen J. Analysis of Aspergillus nidulans metabolism at the genome-scale. BMC Genomics. 2008;9:1–15. doi: 10.1186/1471-2164-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenman H.C., Casadevall A. Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 2012;93:931–940. doi: 10.1007/s00253-011-3777-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist A.M., Palsson B. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat. Biotechnol. 2008;26:659–667. doi: 10.1038/nbt1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumura M., Solano F., Matsunaga N., Sakai C., Spritz R.A., Hearing V.J. Metal ligand-binding specificities of the tyrosinase-related proteins. Biochem. Biophys. Res. Commun. 1998;585:579–585. doi: 10.1006/bbrc.1997.8007. [DOI] [PubMed] [Google Scholar]
- García-Borrón J.C., Solano F. Molecular anatomy of tyrosinase and its related proteins: Beyond the histidine-bound metal catalytic center. Pigment Cell Res. 2002;15:162–173. doi: 10.1034/j.1600-0749.2002.02012.x. [DOI] [PubMed] [Google Scholar]
- Geis P.A., Szaniszlo P.J. Carotenoid pigments of the dematiaceous fungus Wangiella dermatitidis. Mycologia. 1984;76:268–273. doi: 10.1007/BF00410729. [DOI] [PubMed] [Google Scholar]
- Geis P.A. University of Texas at Austin; 1981. Chemical composition of the yeast and sclerotic cell walls of Wangiella dermatitidis. [Google Scholar]
- Kumar A.K., Vatsyayan P. Production of lipid and fatty acids during growth of aspergillus terreus on hydrocarbon substrates. Appl. Biochem. Biotechnol. 2010;160:1293–1300. doi: 10.1007/s12010-009-8669-x. [DOI] [PubMed] [Google Scholar]
- Kumar, J. (2018). Adaptations of Exophiala dermatitidis in stressful environments. Dissertation. ETD collection for University of Nebraska - Lincoln. AAI10792984. https://digital commons.unl.edu/disserations/AAI10792984.
- Lipke P.N., Ovalle Rafael. Cell Wall Architecture in Yeast: New Structure and New Challenges. Journal of Bacteriology. 1998;180:3735–3740. doi: 10.1128/jb.180.15.3735-3740.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Gao Q., Xu N., Liu L. Genome-scale reconstruction and in silico analysis of Aspergillus terreus metabolism. Mol. BioSyst. 2013;9:1939. doi: 10.1039/c3mb70090a. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology Information. (n.d.). www.ncbi.nlm.nih.gov.
- Oberhardt M.A., Palsson B., Papin J.A. Applications of genome-scale metabolic reconstructions. Mol. Syst. Biol. 2009;5:1–15. doi: 10.1038/msb.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos J.S., Agarwala R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- Schmaler-Ripcke J., Sugareva V., Gebhardt P., Winkler R., Kniemeyer O., Heinekamp T., Brakhage A.A. Production of pyomelanin, a second type of melanin, via the tyrosine degradation pathway in Aspergillus fumigatus. Appl. Environ. Microbiol. 2009;75:493–503. doi: 10.1128/AEM.02077-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzler N., Peltroche-Llacsahuanga H., Bestier N., Zundorf J., Lutticken R., Haase G. Effect of melanin and carotenoids of Exophiala (Wangiella) dermatitidis on phagocytosis, oxidative burst, and killing by human neutrophils. Infect. Immun. 1999;67:94–101. doi: 10.1128/iai.67.1.94-101.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch C.L., Sung G.-H., Lopez-Giraldez F., Townsend J.P., Miadlikowska J., Hofstetter V., Gueidan C. The ascomycota tree of life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst. Biol. 2009;58:224–239. doi: 10.1093/sysbio/syp020. [DOI] [PubMed] [Google Scholar]
- Schroeder W.L., Harris S.D., Saha R. Computation-driven analysis of model polyextremo-tolerant fungus exophiala dermatitidis: defensive pigment metabolic costs and human applications. IScience. 2020;23:100980. doi: 10.1016/j.isci.2020.100980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder W.L., Saha R. OptFill: a tool for infeasible cycle-free gapfilling of stoichiometric metabolic models. IScience. 2020;23:100783. doi: 10.1016/j.isci.2019.100783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spritz R.A., Ho L., Furumura M., Hearing V.J. Mutational analysis of copper binding by human tyrosinase. J. Invest. Dermatol. 1997;109:207–212. doi: 10.1111/1523-1747.ep12319351. [DOI] [PubMed] [Google Scholar]
- Strobel I., Breitenbach J., Scheckhuber C.Q., Osiewacz H.D., Sandmann G. Carotenoids and carotenogenic genes in Podospora anserina: Engineering of the carotenoid composition extends the life span of the mycelium. Curr. Genet. 2009;55:175–184. doi: 10.1007/s00294-009-0235-0. [DOI] [PubMed] [Google Scholar]
- Szaniszlo P.J. Molecular genetic studies of the model dematiaceous pathogen Wangiella dermatitidis. Int. J. Med. Microbiol. 2002;292:381–390. doi: 10.1078/1438-4221-00221. [DOI] [PubMed] [Google Scholar]
- Vongsangnak W., Olsen P., Hansen K., Krogsgaard S., Nielsen J. Improved annotation through genome-scale metabolic modeling of Aspergillus oryzae. BMC Genomics. 2008;9:1–14. doi: 10.1186/1471-2164-9-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.S., Lin C.J., Hwang J.K. Prediction of protein subcellular localization. Proteins Struct. Funct. Bioinform. 2006;64:643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- Zhang C., Hua Q. Applications of genome-scale metabolic models in biotechnology and systems medicine. Front. Physiol. 2016;6:1–8. doi: 10.3389/fphys.2015.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets and code generated during this study are available in two GitHub repositories related to this work, one for the Exophiala dermatitidis model iEde2091 (https://doi.org/10.5281/zenodo.3608172) and one for the OptFill tool (https://doi.org/10.5281/zenodo.3518501). These DOIs are for single releases of these GitHub repositories. Repositories which will be updated periodically and can be found through the laboratory group’s GitHub repository pages located at https://github.com/ssbio/E_dermatitidis_model (iEde2091 model and related code) and https://github.com/ssbio/OptFill (OptFill and related code). All data generated in the execution of the protocol has been included in the in appropriate GitHub repository or in the supplemental files provided with the published works associated with this protocol, namely Schroeder and Saha (2020) and Schroeder et al. (2020).