Figure 1.
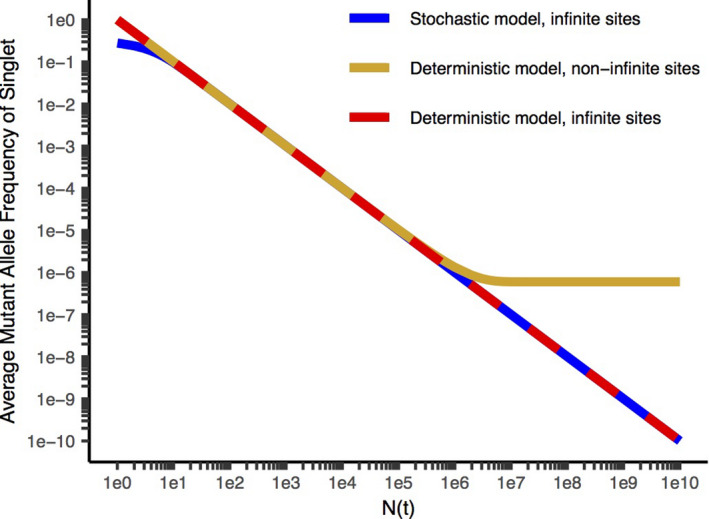
Average MAF (mutant allele frequency) for a given mutation vs. N(t) (the number of cells at the time it is formed), for the stochastic model with the infinite sites assumption 6 (blue), the deterministic model with the infinite sites assumption 5 (red), and without the infinite sites assumption 2 (gold). Sequencing to a depth D queries, on average, mutational events occurring when N(t) = D. Stochastic models consider random fluctuations from the expected average evolutionary trajectory and are more accurate at early times when the tumor is small compared with deterministic models that are based on the expected average evolutionary trajectory. The infinite sites assumption states that a mutation in a particular base will occur uniquely in one cell at any instant. Models without the infinite sites assumption are more accurate than those with it for larger tumor masses in which the number of cells approaches or exceeds the reciprocal of the effective mutation rate, since the expected number of instances of mutations at a particular base is equal to the product of the effective mutation rate and the number of cells dividing. The deterministic model with the infinite sites assumption leads to a reciprocal relationship between N(t) and the average MAF and a straight line with a slope of −1 on a log‐log plot. The deterministic model without infinite sites predicts that the MAF will approach a limiting value equal to the effective mutation rate for N(t) comparable to the reciprocal of the effective mutation rate and larger. This corresponds to a significant degree of additional diversity added late in tumor evolution, including during the clinical course, compared with the other models. Parameters are cell birth rate = 0.25/day, cell death rate = 0.18/day, effective mutation rate per base per new cell added to the tumor = 6.1 × 10−7. Reproduced from ref. 2 with permission.
