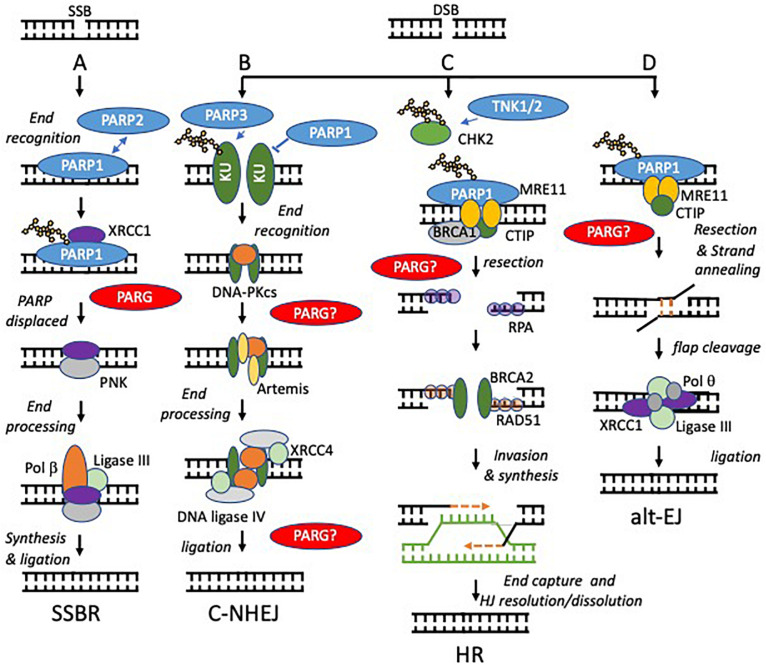
FIGURE 2.
Models of PARP function at single and double strand breaks. Potential sites where PARG function is required to reverse PARylation to facilitate downstream processing are indicated by PARG?, proven roles indicated by PARG. (A) SSBs, arising directly from damage to the DNA backbone or as intermediates in BER, are bound by PARP1 or PARP2. DNA binding activates PARP causing automodification. Automodified PARP recruits XRCC1 to the SSB. PARP is displaced. Damaged 5’- or 3’-termini are processed into 5’-phosphate and 3’-hydroxyl groups by APE1 or PNK. Polβ then performs gap filling followed by ligation by Lig3α. (B,C) PARP activity regulates the relative contribution that non-homologous end-joining (NHEJ) and homologous recombination (HR) make to repair of DNA double strand breaks (DSBs). NHEJ begins with binding of the DNA ends by the Ku70/80 heterodimer, which recruits DNA-PKcs. If the ends are not compatible, they are trimmed by nucleases, e.g., Artemis. The ligation complex XRCC4-DNA Ligase IV-XLF then seals the DSB. In HR, MRE11 resects the break to generate single stranded DNA (ssDNA), which is quickly coated and subsequently replaced by Rad51. The Rad51 nucleoprotein filaments mediate strand invasion of the homologous template. Synthesis of DNA using the sister chromatid is then followed by capture of the second end and holiday junction (HJ) resolution or dissolution leading to DSB repair. PARP1 competes with KU for binding at DSBs and promotes resection by MRE11 (a component of the MRN complex, therefore PARP1 activation favors HR. In contrast PARP3 PARylates KU70/80and limits DNA end resection favoring NHEJ. TNK1/2 PARylates CHK2 to promote HR. (D) PARP1 is required to recruit MRE11 to regions of DNA micro-homology in order to initiate alternative end-joining (alt-EJ), following limited resection and strand annealing, the DNA flaps are cleaved, finally Pol q and the ligation complex XRCC1-ligase III seal the gap.